Gene Page: YWHAZ
Summary ?
| GeneID | 7534 |
| Symbol | YWHAZ |
| Synonyms | 14-3-3-zeta|HEL-S-3|HEL-S-93|HEL4|KCIP-1|YWHAD |
| Description | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta |
| Reference | MIM:601288|HGNC:HGNC:12855|Ensembl:ENSG00000164924|HPRD:03183|Vega:OTTHUMG00000134291 |
| Gene type | protein-coding |
| Map location | 8q23.1 |
| Pascal p-value | 0.031 |
| Sherlock p-value | 0.209 |
| Fetal beta | 0.924 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Nucleus accumbens basal ganglia |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION G2Cdb.human_mGluR5 G2Cdb.human_mitochondria G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 11.6823 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| YWHAZ | chr8 | 101937218 | T | C | NM_001135699 NM_001135700 NM_001135701 NM_001135702 NM_003406 NM_145690 | p.115K>R p.115K>R p.115K>R p.115K>R p.115K>R p.115K>R | missense missense missense missense missense missense | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg06776824 | 8 | 101962949 | YWHAZ | 1.84E-5 | 0.873 | 0.016 | DMG:Wockner_2014 |
| cg06692871 | 8 | 101964133 | YWHAZ | 5.67E-5 | -0.524 | 0.022 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs3105460 | 8 | 101937374 | YWHAZ | ENSG00000164924.13 | 1.093E-6 | 0.01 | 28242 | gtex_brain_ba24 |
| rs34699406 | 8 | 101946380 | YWHAZ | ENSG00000164924.13 | 1.095E-6 | 0.01 | 19236 | gtex_brain_ba24 |
| rs3134353 | 8 | 101947453 | YWHAZ | ENSG00000164924.13 | 1.093E-6 | 0.01 | 18163 | gtex_brain_ba24 |
| rs1470764 | 8 | 101950038 | YWHAZ | ENSG00000164924.13 | 1.093E-6 | 0.01 | 15578 | gtex_brain_ba24 |
| rs3134358 | 8 | 101958433 | YWHAZ | ENSG00000164924.13 | 1.138E-6 | 0.01 | 7183 | gtex_brain_ba24 |
| rs983583 | 8 | 101961910 | YWHAZ | ENSG00000164924.13 | 1.155E-6 | 0.01 | 3706 | gtex_brain_ba24 |
| rs71506350 | 8 | 101962901 | YWHAZ | ENSG00000164924.13 | 1.057E-6 | 0.01 | 2715 | gtex_brain_ba24 |
| rs3100052 | 8 | 101967139 | YWHAZ | ENSG00000164924.13 | 1.194E-6 | 0.01 | -1523 | gtex_brain_ba24 |
| rs7817485 | 8 | 101969528 | YWHAZ | ENSG00000164924.13 | 1.279E-6 | 0.01 | -3912 | gtex_brain_ba24 |
| rs4734500 | 8 | 101970119 | YWHAZ | ENSG00000164924.13 | 8.091E-7 | 0.01 | -4503 | gtex_brain_ba24 |
| rs2386921 | 8 | 101975378 | YWHAZ | ENSG00000164924.13 | 4.478E-7 | 0.01 | -9762 | gtex_brain_ba24 |
| rs2386922 | 8 | 101975479 | YWHAZ | ENSG00000164924.13 | 2.485E-7 | 0.01 | -9863 | gtex_brain_ba24 |
| rs2923781 | 8 | 101976510 | YWHAZ | ENSG00000164924.13 | 7.683E-7 | 0.01 | -10894 | gtex_brain_ba24 |
| rs57408636 | 8 | 101980444 | YWHAZ | ENSG00000164924.13 | 3.22E-7 | 0.01 | -14828 | gtex_brain_ba24 |
Section II. Transcriptome annotation
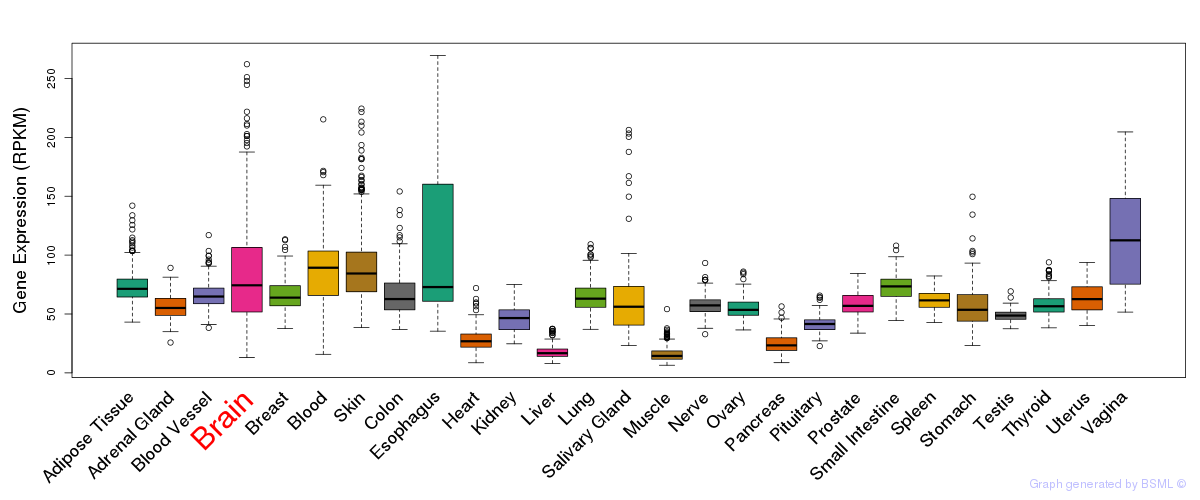
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008134 | transcription factor binding | IPI | 16114898 | |
| GO:0019904 | protein domain specific binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007165 | signal transduction | TAS | 16130169 | |
| GO:0006916 | anti-apoptosis | TAS | 16130169 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | TAS | 16130169 | |
| GO:0042470 | melanosome | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AANAT | AA-NAT | SNAT | arylalkylamine N-acetyltransferase | - | HPRD,BioGRID | 11336675 |
| AANAT | AA-NAT | SNAT | arylalkylamine N-acetyltransferase | AANAT interacts with 14-3-3-zeta isoform. This interaction was modeled on a demonstrated interaction between sheep AANAT and human 14-3-3-zeta. | BIND | 8024705 |11336675 |11427721 |
| ADAM22 | MDC2 | MGC149832 | ADAM metallopeptidase domain 22 | - | HPRD | 12589811 |
| ADRA2A | ADRA2 | ADRA2R | ADRAR | ALPHA2AAR | ZNF32 | adrenergic, alpha-2A-, receptor | - | HPRD | 10224112 |
| ADRA2B | ADRA2L1 | ADRA2RL1 | ADRARL1 | ALPHA2BAR | adrenergic, alpha-2B-, receptor | - | HPRD | 10224112 |
| ADRA2C | ADRA2L2 | ADRA2RL2 | ADRARL2 | ALPHA2CAR | adrenergic, alpha-2C-, receptor | - | HPRD | 10224112 |
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | - | HPRD,BioGRID | 11956222 |
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | Akt interaction with and phosphorylation of 14-3-3 zeta at the same residue as MAPKAPK2. | BIND | 11956222 |
| ARHGEF2 | DKFZp547L106 | DKFZp547P1516 | GEF | GEF-H1 | GEFH1 | KIAA0651 | LFP40 | P40 | rho/rac guanine nucleotide exchange factor (GEF) 2 | - | HPRD | 14970201 |
| BAD | BBC2 | BCL2L8 | BCL2-associated agonist of cell death | - | HPRD,BioGRID | 11410287 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | - | HPRD,BioGRID | 10026197 |
| BCR | ALL | BCR-ABL1 | BCR1 | CML | D22S11 | D22S662 | FLJ16453 | PHL | breakpoint cluster region | - | HPRD | 7939633 |
| BRAF | B-RAF1 | BRAF1 | FLJ95109 | MGC126806 | MGC138284 | RAFB1 | v-raf murine sarcoma viral oncogene homolog B1 | Affinity Capture-MS | BioGRID | 17353931 |
| BRAF | B-RAF1 | BRAF1 | FLJ95109 | MGC126806 | MGC138284 | RAFB1 | v-raf murine sarcoma viral oncogene homolog B1 | - | HPRD | 8668348 |
| BSPRY | FLJ20150 | B-box and SPRY domain containing | - | HPRD | 12615066 |
| C22orf9 | - | chromosome 22 open reading frame 9 | Affinity Capture-MS | BioGRID | 17353931 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | Two-hybrid | BioGRID | 9367879 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | c-Cbl interacts with 14-3-3-zeta. | BIND | 9367879 |
| CDC25A | CDC25A2 | cell division cycle 25 homolog A (S. pombe) | - | HPRD,BioGRID | 11912208 |
| CDC25B | - | cell division cycle 25 homolog B (S. pombe) | - | HPRD,BioGRID | 10713667 |
| CDC25B | - | cell division cycle 25 homolog B (S. pombe) | 14-3-3-zeta interacts with phosphorylated CDC25B. | BIND | 15629715 |
| CDC25C | CDC25 | cell division cycle 25 homolog C (S. pombe) | - | HPRD,BioGRID | 10864927 |
| CLIC4 | CLIC4L | DKFZp566G223 | FLJ38640 | H1 | MTCLIC | huH1 | p64H1 | chloride intracellular channel 4 | - | HPRD | 11563969 |
| CSF2RB | CD131 | CDw131 | IL3RB | IL5RB | colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) | - | HPRD,BioGRID | 8278375 |10477722 |
| DFFA | DFF-45 | DFF1 | ICAD | DNA fragmentation factor, 45kDa, alpha polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
| EDC3 | FLJ21128 | FLJ31777 | LSM16 | YJDC | YJEFN2 | enhancer of mRNA decapping 3 homolog (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | EGFR interacts with 14-3-3-zeta. | BIND | 15225635 |
| EPB41L3 | 4.1B | DAL-1 | DAL1 | FLJ37633 | KIAA0987 | erythrocyte membrane protein band 4.1-like 3 | Affinity Capture-MS | BioGRID | 17353931 |
| FOXO1 | FKH1 | FKHR | FOXO1A | forkhead box O1 | - | HPRD,BioGRID | 11237865 |
| FOXO3 | AF6q21 | DKFZp781A0677 | FKHRL1 | FKHRL1P2 | FOXO2 | FOXO3A | MGC12739 | MGC31925 | forkhead box O3 | - | HPRD,BioGRID | 10102273 |
| GP1BA | BSS | CD42B | CD42b-alpha | GP1B | MGC34595 | glycoprotein Ib (platelet), alpha polypeptide | - | HPRD,BioGRID | 8631758 |9454760 |10627461|9454760 |
| GP1BB | CD42c | glycoprotein Ib (platelet), beta polypeptide | - | HPRD | 8034572 |10627461 |
| GP1BB | CD42c | glycoprotein Ib (platelet), beta polypeptide | in vitro in vivo Two-hybrid | BioGRID | 8034572 |9454760 |10627461 |
| HDAC4 | HA6116 | HD4 | HDAC-A | HDACA | KIAA0288 | histone deacetylase 4 | Reconstituted Complex | BioGRID | 11504882 |
| HDAC4 | HA6116 | HD4 | HDAC-A | HDACA | KIAA0288 | histone deacetylase 4 | - | HPRD | 11486037 |
| HMGN1 | FLJ27265 | FLJ31471 | HMG14 | MGC104230 | MGC117425 | high-mobility group nucleosome binding domain 1 | - | HPRD,BioGRID | 12215538 |
| HNRNPL | FLJ35509 | HNRPL | P/OKcl.14 | hnRNP-L | heterogeneous nuclear ribonucleoprotein L | Affinity Capture-MS | BioGRID | 17353931 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | - | HPRD | 9581554 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | IGFIR interacts with 14-3-3-zeta isoform. | BIND | 9581554 |
| IL9R | CD129 | interleukin 9 receptor | - | HPRD,BioGRID | 10642536 |
| INPP5A | 5PTASE | DKFZp434A1721 | MGC116947 | MGC116949 | inositol polyphosphate-5-phosphatase, 40kDa | - | HPRD,BioGRID | 9398266 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | - | HPRD,BioGRID | 9312143 |
| IRS2 | - | insulin receptor substrate 2 | - | HPRD,BioGRID | 9312143 |
| KIAA1377 | - | KIAA1377 | Two-hybrid | BioGRID | 16169070 |
| KIF1C | KIAA0706 | LTXS1 | kinesin family member 1C | - | HPRD | 10559254 |
| KIF5B | KINH | KNS | KNS1 | UKHC | kinesin family member 5B | Affinity Capture-MS | BioGRID | 17353931 |
| KLC1 | KLC | KNS2 | KNS2A | MGC15245 | kinesin light chain 1 | Affinity Capture-MS | BioGRID | 17353931 |
| KLC4 | KNSL8 | MGC111777 | bA387M24.3 | kinesin light chain 4 | Affinity Capture-MS | BioGRID | 17353931 |
| KRT18 | CYK18 | K18 | keratin 18 | - | HPRD | 9524113 |11917136 |
| LARP1 | KIAA0731 | LARP | MGC19556 | La ribonucleoprotein domain family, member 1 | Affinity Capture-MS | BioGRID | 17353931 |
| LIMK1 | LIMK | LIM domain kinase 1 | - | HPRD,BioGRID | 12323073 |
| LYST | CHS | CHS1 | lysosomal trafficking regulator | Reconstituted Complex | BioGRID | 11984006 |
| MAP3K2 | MEKK2 | MEKK2B | mitogen-activated protein kinase kinase kinase 2 | - | HPRD | 9452471 |
| MAP3K3 | MAPKKK3 | MEKK3 | mitogen-activated protein kinase kinase kinase 3 | - | HPRD | 9452471 |
| MAP3K5 | ASK1 | MAPKKK5 | MEKK5 | mitogen-activated protein kinase kinase kinase 5 | - | HPRD,BioGRID | 10411906|11336675 |
| MAPKAPK2 | MK2 | mitogen-activated protein kinase-activated protein kinase 2 | MAPKAPK2 interacts with and phosphorylates 14-3-3-zeta. | BIND | 12861023 |
| MAPT | DDPAC | FLJ31424 | FTDP-17 | MAPTL | MGC138549 | MSTD | MTBT1 | MTBT2 | PPND | TAU | microtubule-associated protein tau | Tau interacts with 14-3-3-zeta. | BIND | 12176984 |
| MAPT | DDPAC | FLJ31424 | FTDP-17 | MAPTL | MGC138549 | MSTD | MTBT1 | MTBT2 | PPND | TAU | microtubule-associated protein tau | - | HPRD,BioGRID | 10840038 |
| MLF1 | - | myeloid leukemia factor 1 | - | HPRD | 12176995 |
| NADK | FLJ13052 | FLJ37724 | dJ283E3.1 | NAD kinase | Affinity Capture-MS | BioGRID | 17353931 |
| NCKAP1 | FLJ11291 | HEM2 | KIAA0587 | MGC8981 | NAP1 | NAP125 | NCK-associated protein 1 | Affinity Capture-MS | BioGRID | 17353931 |
| NFATC2 | KIAA0611 | NFAT1 | NFATP | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | - | HPRD,BioGRID | 10611249 |
| NFATC4 | NF-ATc4 | NFAT3 | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 | - | HPRD,BioGRID | 10611249 |
| NOLC1 | KIAA0035 | NOPP130 | NOPP140 | NS5ATP13 | P130 | nucleolar and coiled-body phosphoprotein 1 | Affinity Capture-MS | BioGRID | 17353931 |
| PABPC4 | APP-1 | APP1 | FLJ43938 | PABP4 | iPABP | poly(A) binding protein, cytoplasmic 4 (inducible form) | Affinity Capture-MS | BioGRID | 17353931 |
| PAK4 | - | p21 protein (Cdc42/Rac)-activated kinase 4 | Affinity Capture-MS | BioGRID | 17353931 |
| PCTK1 | FLJ16665 | PCTAIRE | PCTAIRE1 | PCTGAIRE | PCTAIRE protein kinase 1 | - | HPRD,BioGRID | 9197417 |
| PDC | MEKA | PHD | PhLOP | PhLP | phosducin | - | HPRD | 11287646 |11331285 |
| PFKFB2 | DKFZp781D2217 | MGC138308 | MGC138310 | PFK-2/FBPase-2 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 | - | HPRD | 12853467 |
| PRKCA | AAG6 | MGC129900 | MGC129901 | PKC-alpha | PKCA | PRKACA | protein kinase C, alpha | - | HPRD,BioGRID | 11950841 |
| PRKCD | MAY1 | MGC49908 | PKCD | nPKC-delta | protein kinase C, delta | Affinity Capture-Western | BioGRID | 11950841 |
| PRKCE | MGC125656 | MGC125657 | PKCE | nPKC-epsilon | protein kinase C, epsilon | Affinity Capture-Western | BioGRID | 11950841 |
| PRKCI | DXS1179E | MGC26534 | PKCI | nPKC-iota | protein kinase C, iota | Reconstituted Complex | BioGRID | 12893243 |
| PRKCZ | PKC-ZETA | PKC2 | protein kinase C, zeta | Biochemical Activity Reconstituted Complex | BioGRID | 10620507 |12893243 |
| PRKD1 | PKC-MU | PKCM | PKD | PRKCM | protein kinase D1 | Reconstituted Complex | BioGRID | 12893243 |
| PTPN13 | DKFZp686J1497 | FAP-1 | PNP1 | PTP-BAS | PTP-BL | PTP1E | PTPL1 | PTPLE | protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) | - | HPRD | 9305890 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | Raf-1 interacts with 14-3-3 zeta. This interaction was modelled on a demonstrated interaction between human Raf-1 and murine 14-3-3-zeta. | BIND | 7559537 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 7628630 |9261098 |10620507 |10887173 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | - | HPRD | 7628630 |7939632 |
| RAI14 | DKFZp564G013 | KIAA1334 | NORPEG | RAI13 | retinoic acid induced 14 | Affinity Capture-MS | BioGRID | 17353931 |
| RALGPS2 | FLJ10244 | FLJ25604 | KIAA0351 | dJ595C2.1 | Ral GEF with PH domain and SH3 binding motif 2 | Affinity Capture-MS | BioGRID | 17353931 |
| REM1 | GD:REM | GES | MGC48669 | REM | RAS (RAD and GEM)-like GTP-binding 1 | - | HPRD,BioGRID | 10441394 |
| RGS3 | C2PA | FLJ20370 | FLJ31516 | FLJ90496 | PDZ-RGS3 | RGP3 | regulator of G-protein signaling 3 | - | HPRD,BioGRID | 11985497 |
| RIN1 | - | Ras and Rab interactor 1 | - | HPRD | 9144171 |
| RPL31 | MGC88191 | ribosomal protein L31 | Affinity Capture-MS | BioGRID | 17353931 |
| RRAD | RAD | RAD1 | REM3 | Ras-related associated with diabetes | Affinity Capture-Western | BioGRID | 10441394 |
| SAMD4B | FLJ10211 | MGC99832 | SMGB | sterile alpha motif domain containing 4B | Affinity Capture-MS | BioGRID | 17353931 |
| SF3B1 | PRP10 | PRPF10 | SAP155 | SF3b155 | splicing factor 3b, subunit 1, 155kDa | Affinity Capture-MS | BioGRID | 17353931 |
| SFRS10 | DKFZp686F18120 | Htra2-beta | SRFS10 | TRA2-BETA | TRA2B | TRAN2B | splicing factor, arginine/serine-rich 10 (transformer 2 homolog, Drosophila) | Affinity Capture-MS | BioGRID | 17353931 |
| SSH1 | FLJ38102 | KIAA1298 | SSH-1 | slingshot homolog 1 (Drosophila) | SSH-1S interacts with 14-3-3-zeta. | BIND | 15660133 |
| SSH1 | FLJ38102 | KIAA1298 | SSH-1 | slingshot homolog 1 (Drosophila) | SSH-1L interacts with 14-3-3-zeta. | BIND | 15660133 |
| SSH2 | KIAA1725 | MGC78588 | SSH-2 | slingshot homolog 2 (Drosophila) | SSH-2S interacts with 14-3-3-zeta. | BIND | 15660133 |
| SSH3 | FLJ10928 | FLJ20515 | SSH-3 | slingshot homolog 3 (Drosophila) | SSH-3S interacts with 14-3-3-zeta. | BIND | 15660133 |
| SYN2 | SYNII | SYNIIa | SYNIIb | synapsin II | Two-hybrid | BioGRID | 10358015 |
| TERT | EST2 | TCS1 | TP2 | TRT | hEST2 | telomerase reverse transcriptase | Two-hybrid | BioGRID | 10835362 |
| TH | TYH | tyrosine hydroxylase | - | HPRD | 11359875 |
| TJP2 | MGC26306 | X104 | ZO-2 | ZO2 | tight junction protein 2 (zona occludens 2) | Affinity Capture-MS | BioGRID | 17353931 |
| TNFAIP3 | A20 | MGC104522 | MGC138687 | MGC138688 | OTUD7C | TNFA1P2 | tumor necrosis factor, alpha-induced protein 3 | - | HPRD,BioGRID | 9299557 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 9620776 |
| TPH1 | MGC119994 | TPH | TPRH | tryptophan hydroxylase 1 | - | HPRD,BioGRID | 8101440 |
| TSC1 | KIAA0243 | LAM | MGC86987 | TSC | tuberous sclerosis 1 | - | HPRD,BioGRID | 12176984 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | - | HPRD | 12176984 |12438239 |12468542 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | - | HPRD,BioGRID | 12176984 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | Tumor suppressor protein tuberin interacts with 14-3-3-zeta isoform. | BIND | 11533041 |12176984 |
| UCP2 | BMIQ4 | SLC25A8 | UCPH | uncoupling protein 2 (mitochondrial, proton carrier) | UCP2 interacts with 14.3.3-zeta. | BIND | 10785390 |
| UCP2 | BMIQ4 | SLC25A8 | UCPH | uncoupling protein 2 (mitochondrial, proton carrier) | - | HPRD,BioGRID | 10785390 |
| UCP3 | SLC25A9 | uncoupling protein 3 (mitochondrial, proton carrier) | UCP3 interacts with 14.3.3-zeta. | BIND | 10785390 |
| UCP3 | SLC25A9 | uncoupling protein 3 (mitochondrial, proton carrier) | - | HPRD,BioGRID | 10785390 |
| USP8 | FLJ34456 | HumORF8 | KIAA0055 | MGC129718 | UBPY | ubiquitin specific peptidase 8 | Affinity Capture-MS | BioGRID | 17353931 |
| VIM | FLJ36605 | vimentin | - | HPRD,BioGRID | 10887173 |
| XRCC6 | CTC75 | CTCBF | G22P1 | KU70 | ML8 | TLAA | X-ray repair complementing defective repair in Chinese hamster cells 6 | Two-hybrid | BioGRID | 16169070 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | Affinity Capture-Western Reconstituted Complex | BioGRID | 10887173 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | - | HPRD | 11336675 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | 14-3-3zeta homodimerizes. | BIND | 12861023 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG PATHOGENIC ESCHERICHIA COLI INFECTION | 59 | 36 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CHREBP2 PATHWAY | 42 | 35 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN CARDIAC MYOCTES | 67 | 54 | All SZGR 2.0 genes in this pathway |
| SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | 51 | 41 | All SZGR 2.0 genes in this pathway |
| ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | 37 | 29 | All SZGR 2.0 genes in this pathway |
| PID GMCSF PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| PID ATR PATHWAY | 39 | 25 | All SZGR 2.0 genes in this pathway |
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| PID NFAT 3PATHWAY | 54 | 47 | All SZGR 2.0 genes in this pathway |
| PID MTOR 4PATHWAY | 69 | 55 | All SZGR 2.0 genes in this pathway |
| PID IGF1 PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| PID FOXO PATHWAY | 49 | 43 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID IL3 PATHWAY | 27 | 19 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID P38 MK2 PATHWAY | 21 | 19 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN NUC PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID A6B1 A6B4 INTEGRIN PATHWAY | 46 | 35 | All SZGR 2.0 genes in this pathway |
| PID INSULIN GLUCOSE PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI AKT PATHWAY | 35 | 30 | All SZGR 2.0 genes in this pathway |
| PID PI3K PLC TRK PATHWAY | 36 | 31 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN A4B1 PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ILS | 107 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME IL 3 5 AND GM CSF SIGNALING | 43 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME RAP1 SIGNALLING | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | 84 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME DESTABILIZATION OF MRNA BY KSRP | 17 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| FRASOR TAMOXIFEN RESPONSE UP | 51 | 36 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS C UP | 170 | 114 | All SZGR 2.0 genes in this pathway |
| OUELLET OVARIAN CANCER INVASIVE VS LMP UP | 117 | 85 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER PAX8 PPARG DN | 45 | 29 | All SZGR 2.0 genes in this pathway |
| LI LUNG CANCER | 41 | 30 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER SOX9 TARGETS IN PROSTATE DEVELOPMENT DN | 45 | 33 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| DEN INTERACT WITH LCA5 | 26 | 21 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q12 Q22 AMPLICON | 132 | 82 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR UP | 151 | 100 | All SZGR 2.0 genes in this pathway |
| WANG TARGETS OF MLL CBP FUSION UP | 44 | 26 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 DN | 163 | 115 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM DN | 141 | 99 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| JI RESPONSE TO FSH DN | 58 | 43 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC UP | 123 | 75 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND MACROPHAGE | 77 | 50 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND ENDOTHELIUM | 66 | 47 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND FIBROBLAST | 132 | 93 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| GUTIERREZ MULTIPLE MYELOMA UP | 35 | 24 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR1 TARGETS UP | 53 | 39 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS UP | 116 | 87 | All SZGR 2.0 genes in this pathway |
| GUILLAUMOND KLF10 TARGETS UP | 51 | 39 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 590 | 597 | 1A,m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-149 | 172 | 178 | m8 | hsa-miR-149brain | UCUGGCUCCGUGUCUUCACUCC |
| miR-155 | 1353 | 1360 | 1A,m8 | hsa-miR-155 | UUAAUGCUAAUCGUGAUAGGGG |
| miR-193 | 296 | 302 | m8 | hsa-miR-193a | AACUGGCCUACAAAGUCCCAG |
| hsa-miR-193b | AACUGGCCCUCAAAGUCCCGCUUU | ||||
| miR-214 | 613 | 619 | m8 | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-30-5p | 85 | 91 | m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-328 | 294 | 300 | m8 | hsa-miR-328brain | CUGGCCCUCUCUGCCCUUCCGU |
| miR-34b | 334 | 341 | 1A,m8 | hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
| miR-433-3p | 916 | 922 | 1A | hsa-miR-433brain | AUCAUGAUGGGCUCCUCGGUGU |
| miR-451 | 582 | 588 | m8 | hsa-miR-451brain | AAACCGUUACCAUUACUGAGUUU |
| miR-544 | 577 | 583 | 1A | hsa-miR-544 | AUUCUGCAUUUUUAGCAAGU |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 426 | 433 | 1A,m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.