Gene Page: ALMS1
Summary ?
| GeneID | 7840 |
| Symbol | ALMS1 |
| Synonyms | ALSS |
| Description | ALMS1, centrosome and basal body associated protein |
| Reference | MIM:606844|HGNC:HGNC:428|Ensembl:ENSG00000116127|HPRD:06023|Vega:OTTHUMG00000152812 |
| Gene type | protein-coding |
| Map location | 2p13 |
| Pascal p-value | 4.003E-6 |
| Sherlock p-value | 1.106E-6 |
| TADA p-value | 0.007 |
| Fetal beta | 1.795 |
| eGene | Myers' cis & trans Meta |
| Support | CompositeSet Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| Expression | Meta-analysis of gene expression | P value: 1.429 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ALMS1 | chr2 | 73717739 | C | T | NM_015120 | p.2884R>* | nonsense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12478346 | chr2 | 73584359 | ALMS1 | 7840 | 1.593E-4 | cis | ||
| rs6720094 | chr2 | 73588075 | ALMS1 | 7840 | 8.237E-6 | cis | ||
| rs7574291 | chr2 | 73594940 | ALMS1 | 7840 | 5.802E-4 | cis | ||
| rs6747145 | chr2 | 73609176 | ALMS1 | 7840 | 4.238E-4 | cis | ||
| rs7598396 | chr2 | 73652627 | ALMS1 | 7840 | 7.426E-6 | cis | ||
| rs6753344 | chr2 | 73653137 | ALMS1 | 7840 | 8.456E-6 | cis | ||
| rs1083922 | chr2 | 73704002 | ALMS1 | 7840 | 2.119E-5 | cis | ||
| rs7607014 | chr2 | 73730173 | ALMS1 | 7840 | 1.293E-6 | cis | ||
| rs7598660 | chr2 | 73740424 | ALMS1 | 7840 | 3.845E-5 | cis | ||
| rs11688718 | chr2 | 73760938 | ALMS1 | 7840 | 2.709E-5 | cis | ||
| rs7566315 | chr2 | 73791807 | ALMS1 | 7840 | 7.532E-5 | cis | ||
| rs10190002 | chr2 | 73794053 | ALMS1 | 7840 | 7.367E-6 | cis | ||
| rs7607892 | chr2 | 73818706 | ALMS1 | 7840 | 2.375E-6 | cis | ||
| rs13391552 | chr2 | 73818935 | ALMS1 | 7840 | 8.631E-7 | cis | ||
| rs6710438 | chr2 | 73820107 | ALMS1 | 7840 | 1.508E-6 | cis | ||
| rs4346412 | chr2 | 73843570 | ALMS1 | 7840 | 1.223E-5 | cis | ||
| rs4852939 | chr2 | 73856768 | ALMS1 | 7840 | 9.194E-6 | cis | ||
| rs4852316 | chr2 | 73919657 | ALMS1 | 7840 | 5.659E-5 | cis | ||
| rs17350056 | chr2 | 73922876 | ALMS1 | 7840 | 5.473E-4 | cis | ||
| rs12052539 | chr2 | 73937152 | ALMS1 | 7840 | 5.883E-5 | cis | ||
| rs1806683 | chr2 | 73939811 | ALMS1 | 7840 | 0 | cis | ||
| rs11894953 | chr2 | 73964630 | ALMS1 | 7840 | 0 | cis | ||
| rs17350188 | chr2 | 73964841 | ALMS1 | 7840 | 9.766E-4 | cis | ||
| rs12624267 | chr2 | 73986930 | ALMS1 | 7840 | 5.481E-4 | cis | ||
| rs12478346 | chr2 | 73584359 | ALMS1 | 7840 | 0.02 | trans | ||
| rs6720094 | chr2 | 73588075 | ALMS1 | 7840 | 0 | trans | ||
| rs7574291 | chr2 | 73594940 | ALMS1 | 7840 | 0.04 | trans | ||
| rs6747145 | chr2 | 73609176 | ALMS1 | 7840 | 0.03 | trans | ||
| rs7598396 | chr2 | 73652627 | ALMS1 | 7840 | 0 | trans | ||
| rs6753344 | chr2 | 73653137 | ALMS1 | 7840 | 0 | trans | ||
| rs1083922 | chr2 | 73704002 | ALMS1 | 7840 | 0 | trans | ||
| rs7607014 | chr2 | 73730173 | ALMS1 | 7840 | 2.353E-4 | trans | ||
| rs7598660 | chr2 | 73740424 | ALMS1 | 7840 | 0 | trans | ||
| rs11688718 | chr2 | 73760938 | ALMS1 | 7840 | 0 | trans | ||
| rs7566315 | chr2 | 73791807 | ALMS1 | 7840 | 0.01 | trans | ||
| rs10190002 | chr2 | 73794053 | ALMS1 | 7840 | 0 | trans | ||
| rs7607892 | chr2 | 73818706 | ALMS1 | 7840 | 4.118E-4 | trans | ||
| rs13391552 | chr2 | 73818935 | ALMS1 | 7840 | 1.631E-4 | trans | ||
| rs6710438 | chr2 | 73820107 | ALMS1 | 7840 | 2.732E-4 | trans | ||
| rs4346412 | chr2 | 73843570 | ALMS1 | 7840 | 0 | trans | ||
| rs4852939 | chr2 | 73856768 | ALMS1 | 7840 | 0 | trans | ||
| rs4852316 | chr2 | 73919657 | ALMS1 | 7840 | 0.01 | trans | ||
| rs17350056 | chr2 | 73922876 | ALMS1 | 7840 | 0.04 | trans | ||
| rs12052539 | chr2 | 73937152 | ALMS1 | 7840 | 0.01 | trans | ||
| rs1806683 | chr2 | 73939811 | ALMS1 | 7840 | 0.09 | trans | ||
| rs17350188 | chr2 | 73964841 | ALMS1 | 7840 | 0.07 | trans | ||
| rs12624267 | chr2 | 73986930 | ALMS1 | 7840 | 0.04 | trans |
Section II. Transcriptome annotation
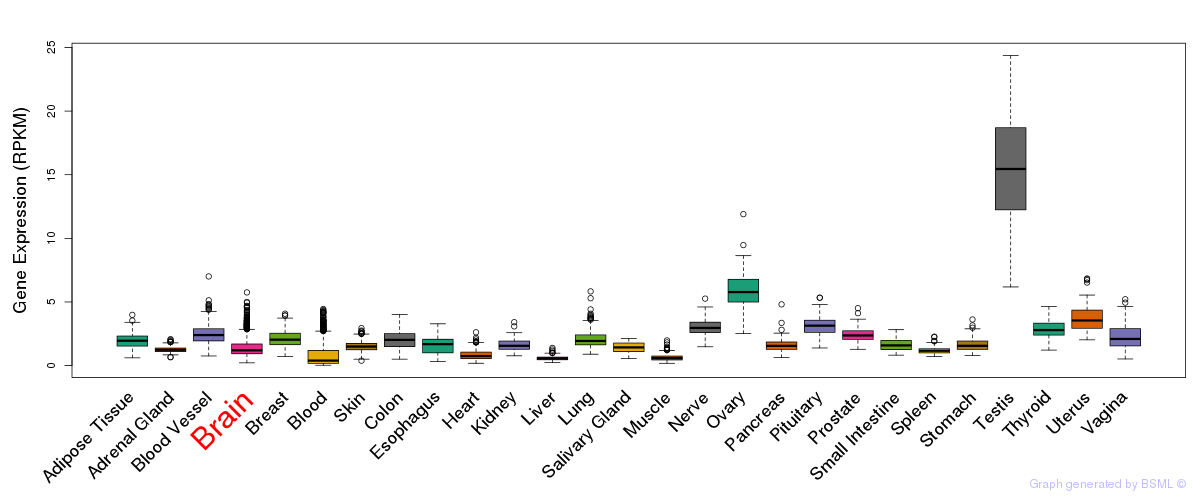
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003674 | molecular_function | ND | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007605 | sensory perception of sound | IEA | - | |
| GO:0007601 | visual perception | IEA | - | |
| GO:0050896 | response to stimulus | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005813 | centrosome | IDA | 15855349 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005929 | cilium | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | 66 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | 59 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G2 G2 M PHASES | 81 | 50 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING DN | 46 | 29 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| CHEBOTAEV GR TARGETS DN | 120 | 73 | All SZGR 2.0 genes in this pathway |
| LIU COMMON CANCER GENES | 79 | 47 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| SU PANCREAS | 54 | 30 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR DN | 160 | 101 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| SPIRA SMOKERS LUNG CANCER DN | 22 | 11 | All SZGR 2.0 genes in this pathway |
| VANASSE BCL2 TARGETS UP | 40 | 25 | All SZGR 2.0 genes in this pathway |
| LEE DIFFERENTIATING T LYMPHOCYTE | 200 | 115 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE DN | 204 | 114 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN DN | 150 | 99 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 DN | 157 | 106 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |