Gene Page: CALR
Summary ?
| GeneID | 811 |
| Symbol | CALR |
| Synonyms | CRT|HEL-S-99n|RO|SSA|cC1qR |
| Description | calreticulin |
| Reference | MIM:109091|HGNC:HGNC:1455|Ensembl:ENSG00000179218|HPRD:00169|Vega:OTTHUMG00000180574 |
| Gene type | protein-coding |
| Map location | 19p13.13 |
| Pascal p-value | 0.041 |
| Sherlock p-value | 0.055 |
| Fetal beta | 0.049 |
| eGene | Myers' cis & trans |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2140065 | chr1 | 241390757 | CALR | 811 | 0.06 | trans |
Section II. Transcriptome annotation
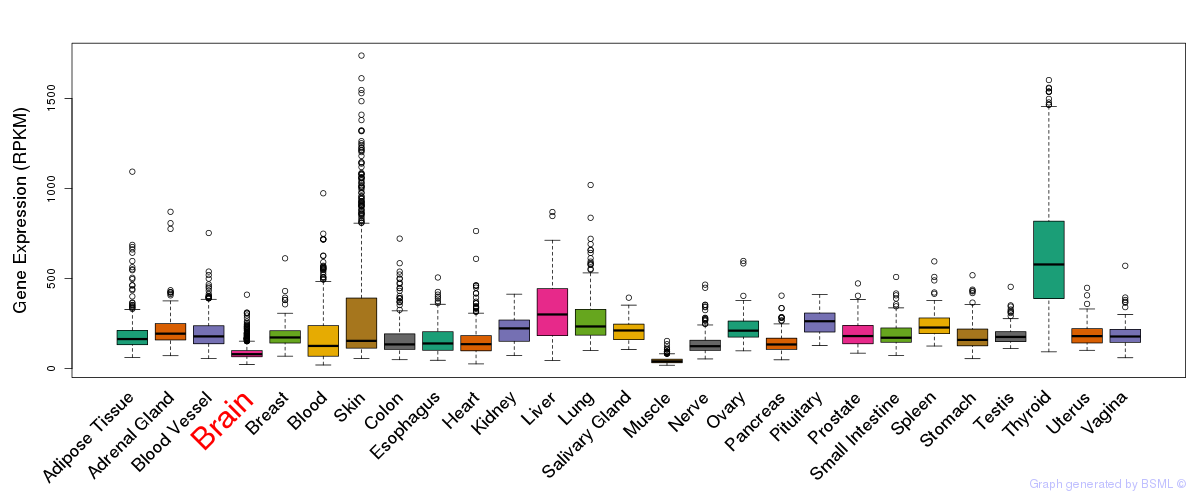
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SYNPO | 0.95 | 0.95 |
| PACSIN1 | 0.95 | 0.93 |
| CAMTA2 | 0.95 | 0.87 |
| MICAL2 | 0.93 | 0.84 |
| MFSD4 | 0.92 | 0.92 |
| DGKZ | 0.92 | 0.88 |
| EXTL1 | 0.92 | 0.92 |
| C8orf46 | 0.91 | 0.89 |
| KCNAB2 | 0.91 | 0.90 |
| SLC17A7 | 0.91 | 0.72 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.60 | -0.71 |
| TRAF4 | -0.54 | -0.67 |
| TUBB2B | -0.54 | -0.63 |
| KIAA1949 | -0.54 | -0.55 |
| PDE9A | -0.53 | -0.63 |
| SH3BP2 | -0.53 | -0.63 |
| SH2B2 | -0.53 | -0.63 |
| DYNLT1 | -0.53 | -0.70 |
| FADS2 | -0.53 | -0.53 |
| PPP1R14B | -0.52 | -0.69 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APOB | FLDB | apolipoprotein B (including Ag(x) antigen) | - | HPRD,BioGRID | 10513896 |12397072 |
| APP | AAA | ABETA | ABPP | AD1 | APPI | CTFgamma | CVAP | PN2 | amyloid beta (A4) precursor protein | - | HPRD,BioGRID | 11378243 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD,BioGRID | 8107809 |
| B2M | - | beta-2-microglobulin | - | HPRD | 8769474 |
| CALR | CRT | FLJ26680 | RO | SSA | cC1qR | calreticulin | CRT interacts with itself to form a dimer. | BIND | 15383281 |
| CANX | CNX | FLJ26570 | IP90 | P90 | calnexin | Affinity Capture-Western | BioGRID | 10436013 |
| CD1D | CD1A | MGC34622 | R3 | CD1d molecule | - | HPRD,BioGRID | 12239218 |
| CD40 | Bp50 | CDW40 | MGC9013 | TNFRSF5 | p50 | CD40 molecule, TNF receptor superfamily member 5 | Affinity Capture-MS | BioGRID | 17353931 |
| CD93 | C1QR1 | C1qR(P) | C1qRP | CDw93 | MXRA4 | dJ737E23.1 | CD93 molecule | - | HPRD | 8062452 |
| ELF3 | EPR-1 | ERT | ESE-1 | ESX | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) | Affinity Capture-MS | BioGRID | 17353931 |
| EPB41 | 4.1R | EL1 | HE | erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) | Affinity Capture-MS | BioGRID | 17353931 |
| F5 | FVL | PCCF | coagulation factor V (proaccelerin, labile factor) | Affinity Capture-Western | BioGRID | 9525969 |
| F8 | AHF | DXS1253E | F8B | F8C | FVIII | HEMA | coagulation factor VIII, procoagulant component | - | HPRD | 9525969 |
| FBN1 | FBN | MASS | MFS1 | OCTD | SGS | WMS | fibrillin 1 | - | HPRD | 10547375 |
| FGB | MGC104327 | MGC120405 | fibrinogen beta chain | - | HPRD,BioGRID | 7592883 |
| HLA-A | HLAA | major histocompatibility complex, class I, A | CRT interacts with HLA-A2. This interaction was modeled on a demonstrated interaction between rabbit CRT and human HLA-A2. | BIND | 15383281 |
| HLA-C | D6S204 | FLJ27082 | HLA-Cw | HLA-Cw12 | HLA-JY3 | HLC-C | PSORS1 | major histocompatibility complex, class I, C | - | HPRD | 8769474 |
| ITGA2B | CD41 | CD41B | GP2B | GPIIb | GTA | HPA3 | integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) | - | HPRD | 7559457 |
| ITGA3 | CD49C | FLJ34631 | FLJ34704 | GAP-B3 | GAPB3 | MSK18 | VCA-2 | VL3A | VLA3a | integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) | - | HPRD | 8006073|11327697 |
| ITGA3 | CD49C | FLJ34631 | FLJ34704 | GAP-B3 | GAPB3 | MSK18 | VCA-2 | VL3A | VLA3a | integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) | - | HPRD | 11327697 |
| LRP1 | A2MR | APOER | APR | CD91 | FLJ16451 | IGFBP3R | LRP | MGC88725 | TGFBR5 | low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) | - | HPRD,BioGRID | 12821648 |14980518 |
| MBL2 | COLEC1 | HSMBPC | MBL | MBP | MBP1 | MGC116832 | MGC116833 | mannose-binding lectin (protein C) 2, soluble (opsonic defect) | - | HPRD,BioGRID | 8062452 |
| NKX2-1 | BCH | BHC | NK-2 | NKX2.1 | NKX2A | TEBP | TITF1 | TTF-1 | TTF1 | NK2 homeobox 1 | - | HPRD,BioGRID | 9988700 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | - | HPRD,BioGRID | 8107808 |9089287 |
| PDIA3 | ER60 | ERp57 | ERp60 | ERp61 | GRP57 | GRP58 | HsT17083 | P58 | PI-PLC | protein disulfide isomerase family A, member 3 | - | HPRD,BioGRID | 10436013 |
| PDIA3 | ER60 | ERp57 | ERp60 | ERp61 | GRP57 | GRP58 | HsT17083 | P58 | PI-PLC | protein disulfide isomerase family A, member 3 | ERp57 interacts with CRT. This interaction was modeled on a demonstrated interaction between ERp57 from an unspecified source and rabbit CRT. | BIND | 12052826 |
| PLAT | DKFZp686I03148 | T-PA | TPA | plasminogen activator, tissue | - | HPRD,BioGRID | 9359841 |
| PRF1 | FLH2 | HPLH2 | MGC65093 | P1 | PFN1 | PFP | perforin 1 (pore forming protein) | - | HPRD,BioGRID | 9671507 |
| PRKCSH | AGE-R2 | G19P1 | PCLD | PLD1 | protein kinase C substrate 80K-H | - | HPRD | 8910335 |
| SEC14L2 | C22orf6 | KIAA1186 | KIAA1658 | MGC65053 | SPF | TAP | TAP1 | SEC14-like 2 (S. cerevisiae) | - | HPRD | 9036970 |
| SLC2A1 | DYT17 | DYT18 | GLUT | GLUT1 | MGC141895 | MGC141896 | PED | solute carrier family 2 (facilitated glucose transporter), member 1 | Reconstituted Complex | BioGRID | 8662691 |
| SLC6A4 | 5-HTT | 5HTT | HTT | OCD1 | SERT | hSERT | solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 | Reconstituted Complex | BioGRID | 10364189 |
| TAPBP | NGS17 | TAPA | TPN | TPSN | tapasin | TAP binding protein (tapasin) | - | HPRD,BioGRID | 8769474 |
| TF | DKFZp781D0156 | PRO1557 | PRO2086 | transferrin | - | HPRD,BioGRID | 9312001 |
| TSHR | CHNG1 | LGR3 | MGC75129 | hTSHR-I | thyroid stimulating hormone receptor | - | HPRD | 12383251 |
| TSHR | CHNG1 | LGR3 | MGC75129 | hTSHR-I | thyroid stimulating hormone receptor | TSHR interacts with the molecular chaperone protein calreticulin. This interaction was modeled on a demonstrated interaction between human TSHR and rabbit calreticulin. | BIND | 12383251 |
| VWF | F8VWF | VWD | von Willebrand factor | Affinity Capture-Western | BioGRID | 10887119 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ANTIGEN PROCESSING AND PRESENTATION | 89 | 65 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NFAT PATHWAY | 56 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | 76 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME ER PHAGOSOME PATHWAY | 61 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME DIABETES PATHWAYS | 133 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | 13 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME UNFOLDED PROTEIN RESPONSE | 80 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME POST TRANSLATIONAL PROTEIN MODIFICATION | 188 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME ASPARAGINE N LINKED GLYCOSYLATION | 81 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME CALNEXIN CALRETICULIN CYCLE | 11 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | 13 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLUENZA LIFE CYCLE | 203 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER UP | 96 | 57 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN UP | 184 | 125 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS UP | 269 | 146 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR UP | 55 | 34 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR DN | 214 | 133 | All SZGR 2.0 genes in this pathway |
| HWANG PROSTATE CANCER MARKERS | 28 | 19 | All SZGR 2.0 genes in this pathway |
| LI LUNG CANCER | 41 | 30 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER | 178 | 108 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT AND CANCER BOX1 DN | 10 | 7 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES CORE NINE CORRELATED | 100 | 68 | All SZGR 2.0 genes in this pathway |
| SUNG METASTASIS STROMA UP | 110 | 70 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH PML RARA FUSION | 77 | 62 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION DN | 187 | 122 | All SZGR 2.0 genes in this pathway |
| RADAEVA RESPONSE TO IFNA1 UP | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT DN | 129 | 86 | All SZGR 2.0 genes in this pathway |
| MA MYELOID DIFFERENTIATION UP | 39 | 29 | All SZGR 2.0 genes in this pathway |
| MUNSHI MULTIPLE MYELOMA UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH3 21 UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| MARCHINI TRABECTEDIN RESISTANCE DN | 49 | 34 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND FIBROBLAST | 132 | 93 | All SZGR 2.0 genes in this pathway |
| PELLICCIOTTA HDAC IN ANTIGEN PRESENTATION UP | 64 | 40 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| HONMA DOCETAXEL RESISTANCE | 34 | 23 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL UP | 125 | 68 | All SZGR 2.0 genes in this pathway |
| CUI GLUCOSE DEPRIVATION | 60 | 44 | All SZGR 2.0 genes in this pathway |
| JIANG TIP30 TARGETS DN | 24 | 14 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 12 | 30 | 20 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP | 181 | 106 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 UP | 140 | 94 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |