Gene Page: RIPK1
Summary ?
| GeneID | 8737 |
| Symbol | RIPK1 |
| Synonyms | RIP|RIP-1|RIP1 |
| Description | receptor interacting serine/threonine kinase 1 |
| Reference | MIM:603453|HGNC:HGNC:10019|Ensembl:ENSG00000137275|HPRD:04583|Vega:OTTHUMG00000014134 |
| Gene type | protein-coding |
| Map location | 6p25.2 |
| Pascal p-value | 0.053 |
| Fetal beta | 0.457 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Frontal Cortex BA9 Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0159 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg12534777 | 6 | 3064469 | RIPK1 | 2.56E-9 | -0.011 | 1.88E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9501885 | chr6 | 2741186 | RIPK1 | 8737 | 0.15 | cis | ||
| rs4864466 | chr4 | 54403975 | RIPK1 | 8737 | 0.16 | trans |
Section II. Transcriptome annotation
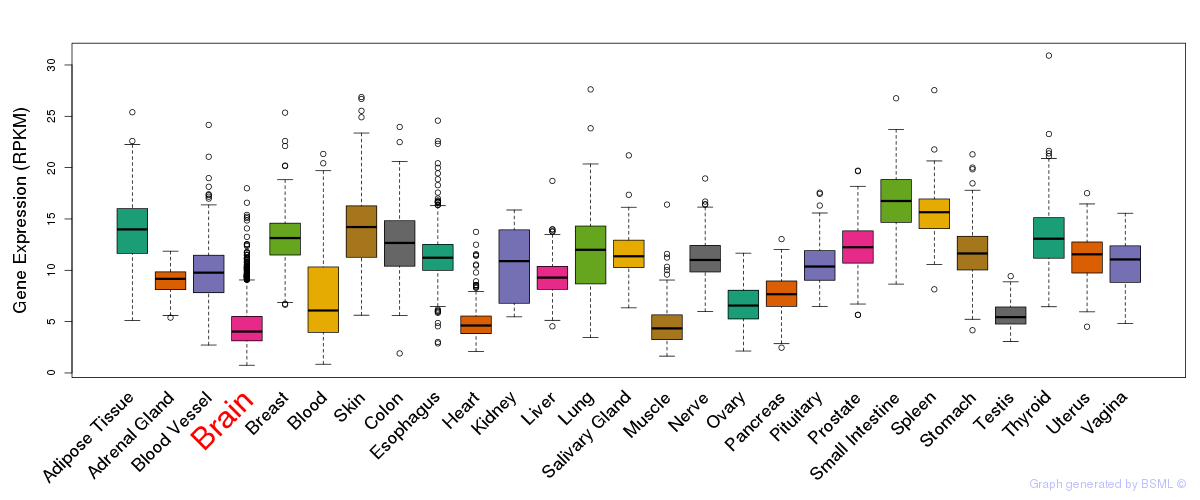
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C3orf49 | 0.77 | 0.77 |
| PLEKHA1 | 0.74 | 0.74 |
| MAPK10 | 0.74 | 0.70 |
| MAN1C1 | 0.74 | 0.77 |
| TBRG1 | 0.73 | 0.75 |
| SMYD2 | 0.73 | 0.72 |
| PIK3R1 | 0.73 | 0.75 |
| IQWD1 | 0.73 | 0.72 |
| SESN1 | 0.73 | 0.71 |
| PIP4K2C | 0.72 | 0.73 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RAB13 | -0.42 | -0.50 |
| CLEC3B | -0.40 | -0.51 |
| AF347015.21 | -0.39 | -0.29 |
| AF347015.18 | -0.37 | -0.27 |
| IL32 | -0.37 | -0.33 |
| BTBD17 | -0.37 | -0.43 |
| GPR39 | -0.35 | -0.39 |
| MPST | -0.35 | -0.34 |
| NME4 | -0.34 | -0.36 |
| APOE | -0.34 | -0.26 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 10356400 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | TAS | 8612133 | |
| GO:0004713 | protein tyrosine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007165 | signal transduction | TAS | 8612133 | |
| GO:0008633 | activation of pro-apoptotic gene products | EXP | 14644197 | |
| GO:0006915 | apoptosis | TAS | 8612133 | |
| GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB cascade | IEP | 12761501 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 7758105 |12692549 |12887920 |15064760 | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CASP10 | ALPS2 | FLICE2 | MCH4 | caspase 10, apoptosis-related cysteine peptidase | Affinity Capture-Western | BioGRID | 11002417 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 11002417 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | - | HPRD,BioGRID | 10837247 |
| CRADD | MGC9163 | RAIDD | CASP2 and RIPK1 domain containing adaptor with death domain | - | HPRD,BioGRID | 8985253 |9044836 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD,BioGRID | 11116146 |
| FADD | GIG3 | MGC8528 | MORT1 | Fas (TNFRSF6)-associated via death domain | Two-hybrid | BioGRID | 11854271 |
| FADD | GIG3 | MGC8528 | MORT1 | Fas (TNFRSF6)-associated via death domain | - | HPRD | 11002422 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | - | HPRD,BioGRID | 7538908 |9740801 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | - | HPRD | 10744744 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | - | HPRD | 14743216 |
| IKBKG | AMCBX1 | FIP-3 | FIP3 | Fip3p | IKK-gamma | IP | IP1 | IP2 | IPD2 | NEMO | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma | - | HPRD,BioGRID | 10880512 |
| MAP3K14 | FTDCR1B | HS | HSNIK | NIK | mitogen-activated protein kinase kinase kinase 14 | - | HPRD,BioGRID | 11116146 |
| MAP3K3 | MAPKKK3 | MEKK3 | mitogen-activated protein kinase kinase kinase 3 | - | HPRD,BioGRID | 11429546 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | FAK interacts with RIP. | BIND | 15121855 |
| RIPK3 | RIP3 | receptor-interacting serine-threonine kinase 3 | - | HPRD,BioGRID | 10339433 |
| RNF216 | TRIAD3 | U7I1 | UBCE7IP1 | ZIN | ring finger protein 216 | - | HPRD,BioGRID | 11854271 |
| SQSTM1 | A170 | OSIL | PDB3 | ZIP3 | p60 | p62 | p62B | sequestosome 1 | - | HPRD,BioGRID | 10356400 |
| TICAM1 | MGC35334 | PRVTIRB | TICAM-1 | TRIF | toll-like receptor adaptor molecule 1 | Affinity Capture-Western | BioGRID | 15064760 |
| TNFAIP3 | A20 | MGC104522 | MGC138687 | MGC138688 | OTUD7C | TNFA1P2 | tumor necrosis factor, alpha-induced protein 3 | A20 ubiquitinates and de-ubiquitinates RIP. | BIND | 15258597 |
| TNFRSF10A | APO2 | CD261 | DR4 | MGC9365 | TRAILR-1 | TRAILR1 | tumor necrosis factor receptor superfamily, member 10a | - | HPRD | 9430227 |
| TNFRSF10B | CD262 | DR5 | KILLER | KILLER/DR5 | TRAIL-R2 | TRAILR2 | TRICK2 | TRICK2A | TRICK2B | TRICKB | ZTNFR9 | tumor necrosis factor receptor superfamily, member 10b | - | HPRD,BioGRID | 9430227 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | - | HPRD,BioGRID | 8612133 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | - | HPRD,BioGRID | 8612133 |
| TRAF1 | EBI6 | MGC:10353 | TNF receptor-associated factor 1 | - | HPRD,BioGRID | 8612133 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | - | HPRD,BioGRID | 8612133 |8702708 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | TRAF2 ubiquitinates RIP. | BIND | 15258597 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | TRAF2 interacts with RIP. | BIND | 8702708 |
| TRAF3 | CAP-1 | CD40bp | CRAF1 | LAP1 | TNF receptor-associated factor 3 | - | HPRD,BioGRID | 8612133 |
| TRAF5 | MGC:39780 | RNF84 | TNF receptor-associated factor 5 | - | HPRD,BioGRID | 11479302 |
| TRIM38 | MGC8946 | RNF15 | RORET | tripartite motif-containing 38 | - | HPRD | 11854271 |
| TRPC4AP | C20orf188 | TRRP4AP | TRUSS | transient receptor potential cation channel, subfamily C, member 4 associated protein | Affinity Capture-Western | BioGRID | 14585990 |
| TUBA3C | TUBA2 | bA408E5.3 | tubulin, alpha 3c | - | HPRD | 14743216 |
| ZFAND5 | ZA20D2 | ZFAND5A | ZNF216 | zinc finger, AN1-type domain 5 | ZNF216 interacts with RIP. | BIND | 14754897 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| KEGG RIG I LIKE RECEPTOR SIGNALING PATHWAY | 71 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG CYTOSOLIC DNA SENSING PATHWAY | 56 | 44 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RELA PATHWAY | 16 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CERAMIDE PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIVNEF PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DEATH PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA KERATINOCYTE PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NFKB PATHWAY | 23 | 19 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P38MAPK PATHWAY | 40 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SODD PATHWAY | 10 | 7 | All SZGR 2.0 genes in this pathway |
| BIOCARTA STRESS PATHWAY | 25 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TNFR1 PATHWAY | 29 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TNFR2 PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| ST TUMOR NECROSIS FACTOR PATHWAY | 29 | 20 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| PID P38 ALPHA BETA PATHWAY | 31 | 25 | All SZGR 2.0 genes in this pathway |
| PID TRAIL PATHWAY | 28 | 20 | All SZGR 2.0 genes in this pathway |
| PID FAS PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| PID TNF PATHWAY | 46 | 33 | All SZGR 2.0 genes in this pathway |
| PID CERAMIDE PATHWAY | 48 | 37 | All SZGR 2.0 genes in this pathway |
| PID HIV NEF PATHWAY | 35 | 26 | All SZGR 2.0 genes in this pathway |
| PID CASPASE PATHWAY | 52 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | 13 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME TRIF MEDIATED TLR3 SIGNALING | 74 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | 18 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB AND MAP KINASES ACTIVATION MEDIATED BY TLR4 SIGNALING REPERTOIRE | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | 10 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING | 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK INTERACTING PROTEINS | 58 | 45 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL UP | 120 | 89 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR UP | 71 | 48 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN | 96 | 71 | All SZGR 2.0 genes in this pathway |
| GENTILE RESPONSE CLUSTER D3 | 61 | 39 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 8 HR UP | 47 | 39 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS EARLY UP | 66 | 44 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR UP | 55 | 41 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |