Gene Page: TNFSF10
Summary ?
| GeneID | 8743 |
| Symbol | TNFSF10 |
| Synonyms | APO2L|Apo-2L|CD253|TL2|TNLG6A|TRAIL |
| Description | tumor necrosis factor superfamily member 10 |
| Reference | MIM:603598|HGNC:HGNC:11925|Ensembl:ENSG00000121858|HPRD:04670|Vega:OTTHUMG00000156917 |
| Gene type | protein-coding |
| Map location | 3q26 |
| Pascal p-value | 0.454 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg16555388 | 3 | 172241216 | TNFSF10 | 0.016 | -5.456 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
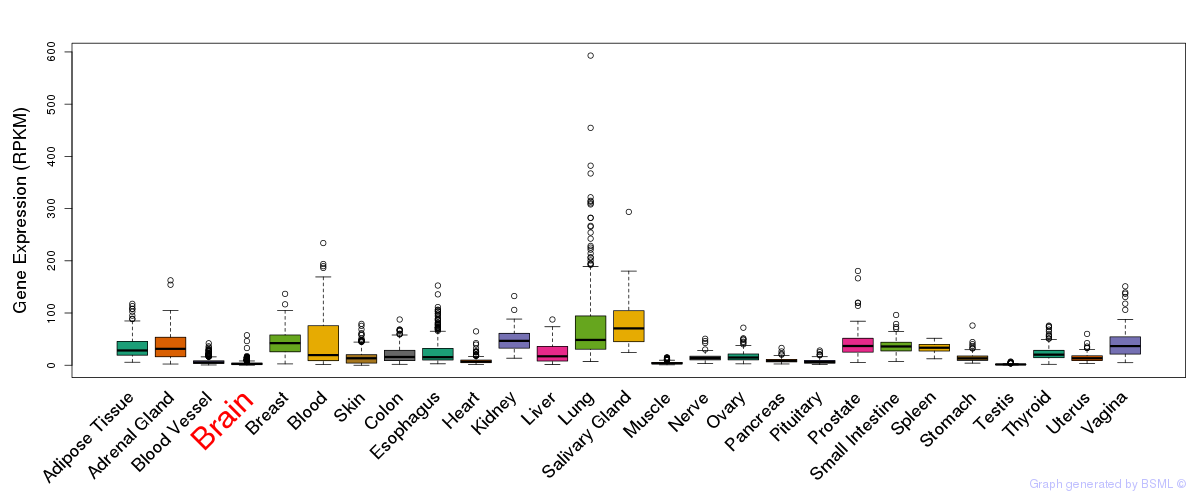
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SAT1 | 0.51 | 0.44 |
| LGALS3 | 0.48 | 0.40 |
| RAB34 | 0.48 | 0.47 |
| SCRG1 | 0.48 | 0.40 |
| DECR1 | 0.46 | 0.44 |
| MT1F | 0.45 | 0.31 |
| MT1G | 0.45 | 0.33 |
| PON3 | 0.44 | 0.44 |
| RTP4 | 0.44 | 0.38 |
| IL17RB | 0.44 | 0.40 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CLCN4 | -0.42 | -0.43 |
| FBXO45 | -0.42 | -0.42 |
| YWHAG | -0.42 | -0.43 |
| TRAPPC9 | -0.42 | -0.41 |
| HECW1 | -0.40 | -0.42 |
| GDI1 | -0.40 | -0.44 |
| DIRAS1 | -0.40 | -0.41 |
| UBQLN2 | -0.40 | -0.39 |
| CNNM2 | -0.40 | -0.39 |
| ADO | -0.40 | -0.40 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACP5 | MGC117378 | TRAP | acid phosphatase 5, tartrate resistant | Two-hybrid | BioGRID | 16169070 |
| C7orf64 | DKFZP564O0523 | DKFZp686D1651 | HSPC304 | chromosome 7 open reading frame 64 | Two-hybrid | BioGRID | 16169070 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | CBP interacts with the TNFSF10 promoter. | BIND | 15619633 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | HDAC1 interacts with the TNFSF10 promoter. | BIND | 15619633 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | HDAC2 interacts with the TNFSF10 promoter. | BIND | 15619633 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | HDAC3 interacts with the TNFSF10 promoter. | BIND | 15619633 |
| HDAC4 | HA6116 | HD4 | HDAC-A | HDACA | KIAA0288 | histone deacetylase 4 | HDAC4 interacts with the TNFSF10 promoter. | BIND | 15619633 |
| HDAC5 | FLJ90614 | HD5 | NY-CO-9 | histone deacetylase 5 | An unspecifed isoform of HDAC5 interacts with the TNFSF10 promoter. | BIND | 15619633 |
| HIST3H3 | H3.4 | H3/g | H3FT | H3t | MGC126886 | MGC126888 | histone cluster 3, H3 | Acetylated H3 interacts with the TNFSF10 promoter. | BIND | 15619633 |
| IER3 | DIF-2 | DIF2 | GLY96 | IEX-1 | IEX-1L | IEX1 | PRG1 | immediate early response 3 | An unspecified isoform of IEX-1 interacts with TNFSF10. | BIND | 15451437 |
| SP1 | - | Sp1 transcription factor | SP1 interacts with the TNFSF10 promoter. | BIND | 15619633 |
| SP3 | DKFZp686O1631 | SPR-2 | Sp3 transcription factor | SP3 interacts with the TNFSF10 promoter. | BIND | 15619633 |
| TNFRSF10A | APO2 | CD261 | DR4 | MGC9365 | TRAILR-1 | TRAILR1 | tumor necrosis factor receptor superfamily, member 10a | - | HPRD,BioGRID | 9082980 |10549288 |
| TNFRSF10B | CD262 | DR5 | KILLER | KILLER/DR5 | TRAIL-R2 | TRAILR2 | TRICK2 | TRICK2A | TRICK2B | TRICKB | ZTNFR9 | tumor necrosis factor receptor superfamily, member 10b | TNFSF10 (TRAIL) interacts with TNFRSF10B (DR5). | BIND | 15688023 |
| TNFRSF10B | CD262 | DR5 | KILLER | KILLER/DR5 | TRAIL-R2 | TRAILR2 | TRICK2 | TRICK2A | TRICK2B | TRICKB | ZTNFR9 | tumor necrosis factor receptor superfamily, member 10b | - | HPRD | 9311998 |10549288 |15110168 |
| TNFRSF10B | CD262 | DR5 | KILLER | KILLER/DR5 | TRAIL-R2 | TRAILR2 | TRICK2 | TRICK2A | TRICK2B | TRICKB | ZTNFR9 | tumor necrosis factor receptor superfamily, member 10b | Affinity Capture-MS Co-crystal Structure Reconstituted Complex | BioGRID | 9311998 |10549288 |11094155 |
| TNFRSF10C | CD263 | DCR1 | LIT | MGC149501 | MGC149502 | TRAILR3 | TRID | tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain | - | HPRD | 9314565 |9613612 |
| TNFRSF10D | CD264 | DCR2 | TRAILR4 | TRUNDD | tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain | - | HPRD,BioGRID | 9430226 |
| TNFRSF11B | MGC29565 | OCIF | OPG | TR1 | tumor necrosis factor receptor superfamily, member 11b | - | HPRD,BioGRID | 9520411 |12221720 |
| TNFSF10 | APO2L | Apo-2L | CD253 | TL2 | TRAIL | tumor necrosis factor (ligand) superfamily, member 10 | - | HPRD,BioGRID | 10651627 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG NATURAL KILLER CELL MEDIATED CYTOTOXICITY | 137 | 92 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DEATH PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| PID TRAIL PATHWAY | 28 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | 13 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS UP | 137 | 94 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| WILCOX RESPONSE TO PROGESTERONE DN | 66 | 44 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| GRABARCZYK BCL11B TARGETS UP | 81 | 40 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D UP | 175 | 108 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL DN | 86 | 59 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT DN | 95 | 57 | All SZGR 2.0 genes in this pathway |
| GRAHAM NORMAL QUIESCENT VS NORMAL DIVIDING UP | 66 | 47 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP UP | 265 | 158 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| HAHTOLA SEZARY SYNDROM UP | 98 | 58 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN DN | 27 | 20 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR DN | 75 | 50 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| OUELLET OVARIAN CANCER INVASIVE VS LMP UP | 117 | 85 | All SZGR 2.0 genes in this pathway |
| BARIS THYROID CANCER DN | 59 | 45 | All SZGR 2.0 genes in this pathway |
| STREICHER LSM1 TARGETS UP | 44 | 34 | All SZGR 2.0 genes in this pathway |
| MARKS HDAC TARGETS UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| DUTTA APOPTOSIS VIA NFKB | 33 | 25 | All SZGR 2.0 genes in this pathway |
| DAUER STAT3 TARGETS DN | 50 | 34 | All SZGR 2.0 genes in this pathway |
| MAINA VHL TARGETS DN | 18 | 14 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| TOMLINS METASTASIS DN | 20 | 16 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP | 223 | 140 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| RADAEVA RESPONSE TO IFNA1 UP | 52 | 40 | All SZGR 2.0 genes in this pathway |
| BROCKE APOPTOSIS REVERSED BY IL6 | 144 | 98 | All SZGR 2.0 genes in this pathway |
| SANA RESPONSE TO IFNG UP | 78 | 50 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS DN | 87 | 69 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE UP | 157 | 105 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR UP | 78 | 56 | All SZGR 2.0 genes in this pathway |
| XU RESPONSE TO TRETINOIN AND NSC682994 UP | 17 | 13 | All SZGR 2.0 genes in this pathway |
| TAVOR CEBPA TARGETS DN | 30 | 21 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP | 183 | 111 | All SZGR 2.0 genes in this pathway |
| BENNETT SYSTEMIC LUPUS ERYTHEMATOSUS | 31 | 21 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA HP UP | 49 | 26 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C8 | 72 | 56 | All SZGR 2.0 genes in this pathway |
| LIANG SILENCED BY METHYLATION 2 | 53 | 34 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS 6HR | 59 | 38 | All SZGR 2.0 genes in this pathway |
| CHEN LVAD SUPPORT OF FAILING HEART DN | 42 | 32 | All SZGR 2.0 genes in this pathway |
| JIANG VHL TARGETS | 138 | 91 | All SZGR 2.0 genes in this pathway |
| BROWNE INTERFERON RESPONSIVE GENES | 68 | 44 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 8HR UP | 105 | 73 | All SZGR 2.0 genes in this pathway |
| MEDINA SMARCA4 TARGETS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS | 108 | 71 | All SZGR 2.0 genes in this pathway |
| GEORGANTAS HSC MARKERS | 71 | 47 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS 3HR | 74 | 47 | All SZGR 2.0 genes in this pathway |
| GAVIN PDE3B TARGETS | 22 | 15 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| LEE METASTASIS AND ALTERNATIVE SPLICING DN | 45 | 31 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING UP | 115 | 80 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| MOSERLE IFNA RESPONSE | 31 | 19 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| BOHN PRIMARY IMMUNODEFICIENCY SYNDROM DN | 40 | 31 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS UP | 153 | 107 | All SZGR 2.0 genes in this pathway |
| LEE EARLY T LYMPHOCYTE DN | 57 | 36 | All SZGR 2.0 genes in this pathway |
| LEE RECENT THYMIC EMIGRANT | 227 | 128 | All SZGR 2.0 genes in this pathway |
| LIU VAV3 PROSTATE CARCINOGENESIS UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G6 DN | 19 | 13 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 LCP WITH H3K4ME3 | 162 | 80 | All SZGR 2.0 genes in this pathway |
| NIELSEN GIST VS SYNOVIAL SARCOMA DN | 20 | 17 | All SZGR 2.0 genes in this pathway |
| WANG METASTASIS OF BREAST CANCER | 15 | 8 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| SUMI HNF4A TARGETS | 34 | 14 | All SZGR 2.0 genes in this pathway |
| BOSCO INTERFERON INDUCED ANTIVIRAL MODULE | 78 | 48 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL DN | 214 | 124 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |