Gene Page: ARHGEF7
Summary ?
| GeneID | 8874 |
| Symbol | ARHGEF7 |
| Synonyms | BETA-PIX|COOL-1|COOL1|Nbla10314|P50|P50BP|P85|P85COOL1|P85SPR|PAK3|PIXB |
| Description | Rho guanine nucleotide exchange factor 7 |
| Reference | MIM:605477|HGNC:HGNC:15607|Ensembl:ENSG00000102606|HPRD:05688|Vega:OTTHUMG00000017357 |
| Gene type | protein-coding |
| Map location | 13q34 |
| Pascal p-value | 0.246 |
| Sherlock p-value | 0.414 |
| Fetal beta | 0.355 |
| DMG | 2 (# studies) |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Nucleus accumbens basal ganglia |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL CompositeSet Darnell FMRP targets Ascano FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14299931 | 13 | 111839335 | ARHGEF7 | 5.177E-4 | 0.577 | 0.047 | DMG:Wockner_2014 |
| cg00557354 | 13 | 111767899 | ARHGEF7 | -0.024 | 0.5 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
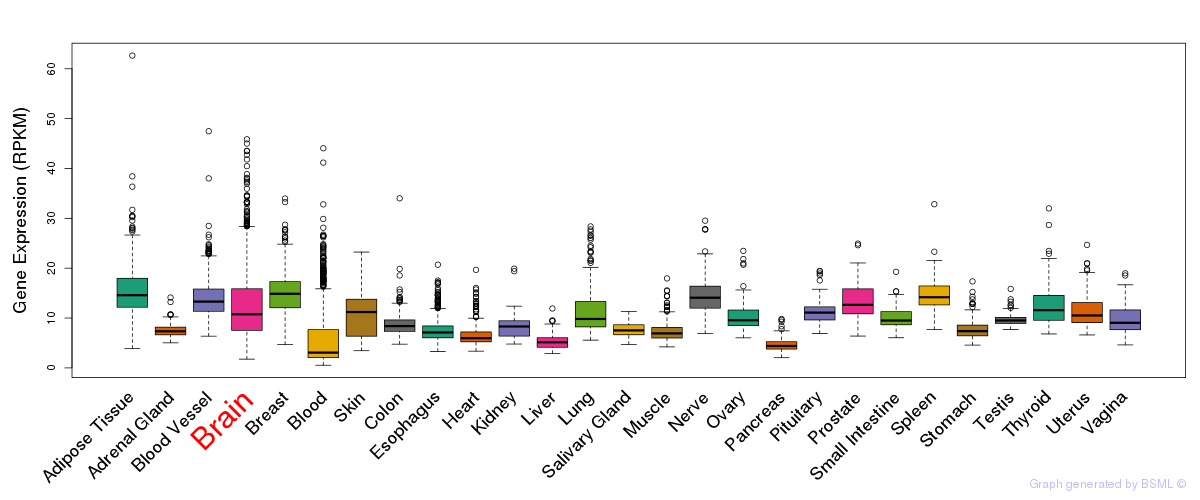
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| YWHAH | 0.82 | 0.84 |
| AC012652.1 | 0.82 | 0.84 |
| RAPGEFL1 | 0.80 | 0.82 |
| PPFIA3 | 0.80 | 0.82 |
| CHGB | 0.80 | 0.82 |
| DCTN1 | 0.79 | 0.79 |
| SGSM1 | 0.79 | 0.82 |
| APLP2 | 0.79 | 0.79 |
| OXCT1 | 0.79 | 0.81 |
| SNCA | 0.79 | 0.82 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| IMPA2 | -0.43 | -0.47 |
| AF347015.21 | -0.41 | -0.20 |
| C21orf57 | -0.40 | -0.38 |
| C1orf61 | -0.38 | -0.33 |
| AC098691.2 | -0.38 | -0.33 |
| RPL35 | -0.38 | -0.40 |
| UXT | -0.37 | -0.32 |
| RPL24 | -0.37 | -0.34 |
| RPS7 | -0.37 | -0.32 |
| C1orf54 | -0.37 | -0.23 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARHGEF6 | COOL2 | Cool-2 | KIAA0006 | MRX46 | PIXA | alpha-PIX | alphaPIX | Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 | ARHGEF6 interacts with ARHGEF7 | BIND | 12499396 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | Two-hybrid | BioGRID | 12935897 |
| CDC42 | CDC42Hs | G25K | cell division cycle 42 (GTP binding protein, 25kDa) | ARHGEF7 (p85Cool-1) interacts with Cdc42. | BIND | 15649357 |
| GIT1 | - | G protein-coupled receptor kinase interacting ArfGAP 1 | Affinity Capture-Western Two-hybrid | BioGRID | 10428811 |12473661 |
| GIT1 | - | G protein-coupled receptor kinase interacting ArfGAP 1 | - | HPRD | 10896954 |
| GIT2 | CAT-2 | DKFZp686G01261 | KIAA0148 | MGC760 | G protein-coupled receptor kinase interacting ArfGAP 2 | - | HPRD | 10428811 |10896954 |
| PAK1 | MGC130000 | MGC130001 | PAKalpha | p21 protein (Cdc42/Rac)-activated kinase 1 | - | HPRD,BioGRID | 11157752|12006652 |
| PAK2 | PAK65 | PAKgamma | p21 protein (Cdc42/Rac)-activated kinase 2 | - | HPRD,BioGRID | 12226077 |
| PAK3 | CDKN1A | MRX30 | MRX47 | OPHN3 | PAK3beta | bPAK | hPAK3 | p21 protein (Cdc42/Rac)-activated kinase 3 | - | HPRD,BioGRID | 9726964 |
| PPM1E | DKFZp781F1422 | KIAA1072 | POPX1 | PP2CH | protein phosphatase 1E (PP2C domain containing) | Reconstituted Complex | BioGRID | 11864573 |
| PPM1F | CaMKPase | FEM-2 | KIAA0015 | POPX2 | hFEM-2 | protein phosphatase 1F (PP2C domain containing) | - | HPRD,BioGRID | 11864573 |
| SHANK1 | SPANK-1 | SSTRIP | synamon | SH3 and multiple ankyrin repeat domains 1 | - | HPRD,BioGRID | 12626503 |
| SHANK2 | CORTBP1 | CTTNBP1 | ProSAP1 | SHANK | SPANK-3 | SH3 and multiple ankyrin repeat domains 2 | - | HPRD,BioGRID | 12626503 |
| SHANK3 | KIAA1650 | PROSAP2 | PSAP2 | SPANK-2 | SH3 and multiple ankyrin repeat domains 3 | Affinity Capture-Western Two-hybrid | BioGRID | 12626503 |
| TGFBR2 | AAT3 | FAA3 | LDS1B | LDS2B | MFS2 | RIIC | TAAD2 | TGFR-2 | TGFbeta-RII | transforming growth factor, beta receptor II (70/80kDa) | T-beta-RII interacts with Beta-PIX. This interaction was modeled on a demonstrated interaction between human T-beta-RII and mouse Beta-PIX. | BIND | 15761153 |
| TUBB3 | MC1R | TUBB4 | beta-4 | tubulin, beta 3 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MYOSIN PATHWAY | 31 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PAR1 PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| ST INTEGRIN SIGNALING PATHWAY | 82 | 62 | All SZGR 2.0 genes in this pathway |
| PID ILK PATHWAY | 45 | 32 | All SZGR 2.0 genes in this pathway |
| PID CDC42 PATHWAY | 70 | 51 | All SZGR 2.0 genes in this pathway |
| PID CDC42 REG PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 INTERNALIZATION PATHWAY | 41 | 35 | All SZGR 2.0 genes in this pathway |
| PID RAC1 REG PATHWAY | 38 | 25 | All SZGR 2.0 genes in this pathway |
| PID AURORA A PATHWAY | 31 | 20 | All SZGR 2.0 genes in this pathway |
| PID FAK PATHWAY | 59 | 45 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA DN | 77 | 52 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| ODONNELL METASTASIS UP | 82 | 58 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA COPY NUMBER DN | 54 | 37 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRACX DN | 20 | 14 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC DN | 123 | 86 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G6 | 153 | 112 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D8 | 40 | 29 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| LIN NPAS4 TARGETS UP | 163 | 100 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE DN | 181 | 97 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| STAMBOLSKY TARGETS OF MUTATED TP53 UP | 49 | 35 | All SZGR 2.0 genes in this pathway |
| STAMBOLSKY RESPONSE TO VITAMIN D3 UP | 84 | 48 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |