Gene Page: BSN
Summary ?
| GeneID | 8927 |
| Symbol | BSN |
| Synonyms | ZNF231 |
| Description | bassoon presynaptic cytomatrix protein |
| Reference | MIM:604020|HGNC:HGNC:1117|Ensembl:ENSG00000164061|HPRD:04933|Vega:OTTHUMG00000133750 |
| Gene type | protein-coding |
| Map location | 3p21.31 |
| Pascal p-value | 0.064 |
| Sherlock p-value | 0.373 |
| Fetal beta | -1.2 |
| DMG | 1 (# studies) |
| Support | PROTEIN CLUSTERING G2Cdb.human_mGluR5 G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets Ascano FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| DNM:Awadalla_2010 | Whole Exome Sequencing analysis | A survey of ~430 Mb of DNA from 401 synapse-expressed genes across all cases and 25 Mb of DNA in controls found 28 candidate DNMs. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BSN | chr3 | 49691984 | G | C | BSN:NM_003458:exon5:c.G4995C:p.V1665V | V1665V | synonymous SNV | Schizophrenia | DNM:Awadalla_2010 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08226300 | 3 | 49695806 | BSN | 2.471E-4 | 0.43 | 0.037 | DMG:Wockner_2014 |
| cg20948091 | 3 | 49608040 | BSN | 2.997E-4 | 0.243 | 0.04 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
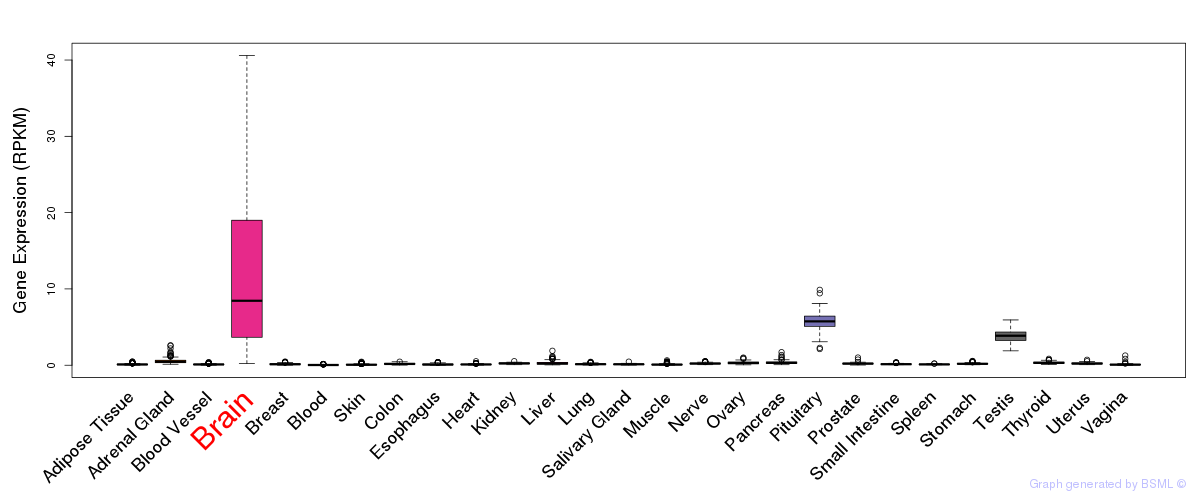
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SH2B2 | 0.75 | 0.69 |
| MARCKSL1 | 0.74 | 0.64 |
| PCGF2 | 0.73 | 0.59 |
| ZBTB12 | 0.73 | 0.62 |
| SH3BP2 | 0.73 | 0.71 |
| SF3A2 | 0.72 | 0.62 |
| FAM110A | 0.72 | 0.55 |
| PKN1 | 0.72 | 0.65 |
| TRAF4 | 0.72 | 0.66 |
| JUP | 0.72 | 0.61 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CCNI2 | -0.60 | -0.66 |
| ITM2B | -0.59 | -0.64 |
| PLA2G4A | -0.58 | -0.66 |
| ME1 | -0.58 | -0.62 |
| ANXA7 | -0.58 | -0.60 |
| OMG | -0.58 | -0.65 |
| CHN1 | -0.57 | -0.66 |
| CACNA1F | -0.57 | -0.66 |
| ACOT13 | -0.56 | -0.63 |
| C5orf53 | -0.56 | -0.60 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 10329005 |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0019717 | synaptosome | IEA | Synap, Brain (GO term level: 7) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005634 | nucleus | TAS | 9806829 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| STARK HYPPOCAMPUS 22Q11 DELETION UP | 53 | 40 | All SZGR 2.0 genes in this pathway |
| STARK BRAIN 22Q11 DELETION | 13 | 10 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| LEIN LOCALIZED TO PROXIMAL DENDRITES | 37 | 26 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 527 | 534 | 1A,m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-124.1 | 871 | 877 | 1A | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-125/351 | 2491 | 2497 | m8 | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-132/212 | 1387 | 1393 | 1A | hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC |
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| miR-135 | 1160 | 1166 | m8 | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-137 | 1058 | 1065 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-143 | 1342 | 1348 | m8 | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
| miR-145 | 1451 | 1457 | m8 | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-150 | 622 | 629 | 1A,m8 | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| miR-153 | 1356 | 1363 | 1A,m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-19 | 1619 | 1625 | m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-218 | 1086 | 1092 | 1A | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-24 | 1950 | 1957 | 1A,m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-25/32/92/363/367 | 3911 | 3917 | m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-30-5p | 3897 | 3903 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG | ||||
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-330 | 486 | 492 | 1A | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-339 | 1037 | 1043 | 1A | hsa-miR-339 | UCCCUGUCCUCCAGGAGCUCA |
| miR-342 | 3970 | 3976 | 1A | hsa-miR-342brain | UCUCACACAGAAAUCGCACCCGUC |
| miR-448 | 1357 | 1363 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-503 | 1559 | 1565 | m8 | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
| miR-539 | 2487 | 2494 | 1A,m8 | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
| miR-9 | 131 | 137 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.