Gene Page: ACVR1
Summary ?
| GeneID | 90 |
| Symbol | ACVR1 |
| Synonyms | ACTRI|ACVR1A|ACVRLK2|ALK2|FOP|SKR1|TSRI |
| Description | activin A receptor type 1 |
| Reference | MIM:102576|HGNC:HGNC:171|Ensembl:ENSG00000115170|HPRD:00021|Vega:OTTHUMG00000131967 |
| Gene type | protein-coding |
| Map location | 2q23-q24 |
| Pascal p-value | 0.326 |
| Sherlock p-value | 0.008 |
| Fetal beta | 0.774 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.02395 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
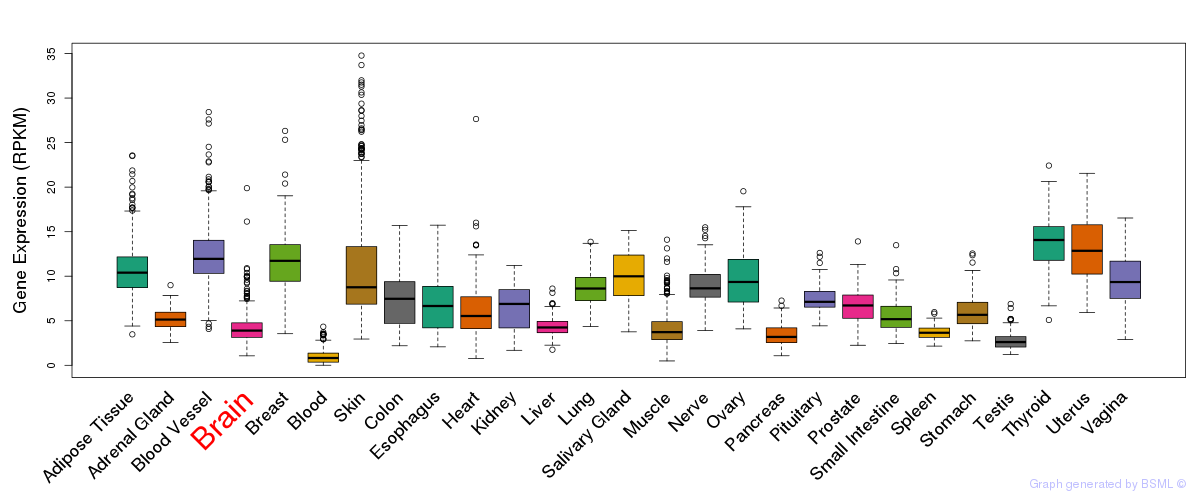
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RAD54L2 | 0.94 | 0.93 |
| CELSR3 | 0.93 | 0.92 |
| KIAA1549 | 0.93 | 0.92 |
| USP49 | 0.93 | 0.92 |
| DCX | 0.93 | 0.90 |
| IGF1R | 0.93 | 0.93 |
| ZMIZ1 | 0.92 | 0.91 |
| BEND3 | 0.92 | 0.91 |
| C20orf117 | 0.91 | 0.92 |
| PLXNA2 | 0.91 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.61 | -0.76 |
| HLA-F | -0.59 | -0.74 |
| AIFM3 | -0.59 | -0.74 |
| SLC16A11 | -0.59 | -0.64 |
| ALDOC | -0.59 | -0.70 |
| SERPINB6 | -0.58 | -0.70 |
| LHPP | -0.58 | -0.59 |
| PTH1R | -0.58 | -0.71 |
| HEPN1 | -0.58 | -0.70 |
| FBXO2 | -0.58 | -0.62 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005524 | ATP binding | IDA | 12065756 | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0016361 | activin receptor activity, type I | IDA | 8242742 | |
| GO:0042803 | protein homodimerization activity | IDA | 18436533 | |
| GO:0030145 | manganese ion binding | IEA | - | |
| GO:0048184 | follistatin binding | NAS | 12702211 | |
| GO:0048185 | activin binding | IDA | 8242742 | |
| GO:0046332 | SMAD binding | IDA | 12065756 | |
| GO:0050431 | transforming growth factor beta binding | IDA | 8242742 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000082 | G1/S transition of mitotic cell cycle | IMP | 9884026 | |
| GO:0001755 | neural crest cell migration | IEA | - | |
| GO:0001569 | patterning of blood vessels | IEA | - | |
| GO:0001701 | in utero embryonic development | IEA | - | |
| GO:0001702 | gastrulation with mouth forming second | IEA | - | |
| GO:0001707 | mesoderm formation | IEA | - | |
| GO:0002526 | acute inflammatory response | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | IDA | 12065756 | |
| GO:0007368 | determination of left/right symmetry | IEA | - | |
| GO:0007281 | germ cell development | IEA | - | |
| GO:0007179 | transforming growth factor beta receptor signaling pathway | IDA | 8242742 | |
| GO:0009790 | embryonic development | IEA | - | |
| GO:0007507 | heart development | IEA | - | |
| GO:0007498 | mesoderm development | IEA | - | |
| GO:0043066 | negative regulation of apoptosis | IMP | 9884026 | |
| GO:0060037 | pharyngeal system development | IEA | - | |
| GO:0030501 | positive regulation of bone mineralization | IMP | 18436533 | |
| GO:0030509 | BMP signaling pathway | IDA | 18436533 | |
| GO:0051145 | smooth muscle cell differentiation | IEA | - | |
| GO:0030278 | regulation of ossification | IMP | 16642017 | |
| GO:0032926 | negative regulation of activin receptor signaling pathway | IMP | 9884026 | |
| GO:0045941 | positive regulation of transcription | IDA | 8242742 | |
| GO:0045669 | positive regulation of osteoblast differentiation | IMP | 18436533 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0045177 | apical part of cell | IEA | - | |
| GO:0048179 | activin receptor complex | IDA | 8242742 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACVR1B | ACTRIB | ACVRLK4 | ALK4 | SKR2 | activin A receptor, type IB | - | HPRD,BioGRID | 8612709 |
| BMP2 | BMP2A | bone morphogenetic protein 2 | - | HPRD | 8006002 |
| BMP7 | OP-1 | bone morphogenetic protein 7 | - | HPRD,BioGRID | 8006002 |
| BMPR2 | BMPR-II | BMPR3 | BMR2 | BRK-3 | FLJ41585 | FLJ76945 | PPH1 | T-ALK | bone morphogenetic protein receptor, type II (serine/threonine kinase) | Affinity Capture-Western Two-hybrid | BioGRID | 7791754 |
| ENG | CD105 | END | FLJ41744 | HHT1 | ORW | ORW1 | endoglin | - | HPRD,BioGRID | 9872992 |
| FNTA | FPTA | MGC99680 | PGGT1A | PTAR2 | farnesyltransferase, CAAX box, alpha | - | HPRD,BioGRID | 8599089 |
| GDF5 | BMP14 | CDMP1 | LAP4 | SYNS2 | growth differentiation factor 5 | - | HPRD,BioGRID | 8702914 |
| IGSF1 | IGCD1 | IGDC1 | INHBP | KIAA0364 | MGC75490 | PGSF2 | immunoglobulin superfamily, member 1 | - | HPRD | 11266516 |
| INHBA | EDF | FRP | inhibin, beta A | - | HPRD | 11266516 |
| INHBB | MGC157939 | inhibin, beta B | - | HPRD | 11266516 |
| INHBC | IHBC | inhibin, beta C | - | HPRD | 12651901 |
| SMAD1 | BSP1 | JV4-1 | JV41 | MADH1 | MADR1 | SMAD family member 1 | - | HPRD | 9748228 |10583507 |10652350 |
| SMAD1 | BSP1 | JV4-1 | JV41 | MADH1 | MADR1 | SMAD family member 1 | Affinity Capture-Western | BioGRID | 9748228 |10652350 |
| SMAD5 | DKFZp781C1895 | DKFZp781O1323 | Dwfc | JV5-1 | MADH5 | SMAD family member 5 | Affinity Capture-Western | BioGRID | 9748228 |10652350 |
| SMAD5 | DKFZp781C1895 | DKFZp781O1323 | Dwfc | JV5-1 | MADH5 | SMAD family member 5 | - | HPRD | 9748228 |10583507 |10652350 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ALK PATHWAY | 37 | 29 | All SZGR 2.0 genes in this pathway |
| PID ALK1 PATHWAY | 26 | 21 | All SZGR 2.0 genes in this pathway |
| PID ALK2 PATHWAY | 11 | 9 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER DN | 169 | 118 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS TURQUOISE UP | 79 | 50 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION UP | 140 | 83 | All SZGR 2.0 genes in this pathway |
| DORSEY GAB2 TARGETS | 31 | 17 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| SARTIPY BLUNTED BY INSULIN RESISTANCE DN | 18 | 14 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 2D UP | 69 | 46 | All SZGR 2.0 genes in this pathway |
| RAY TUMORIGENESIS BY ERBB2 CDC25A UP | 104 | 57 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE DN | 138 | 99 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE EARLY LATE | 317 | 190 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR1 TARGETS UP | 53 | 39 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS UP | 116 | 87 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 568 | 574 | 1A | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-128 | 191 | 197 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-130/301 | 419 | 426 | 1A,m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU | ||||
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-137 | 390 | 396 | 1A | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-140 | 167 | 173 | m8 | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-148/152 | 449 | 455 | m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-182 | 481 | 488 | 1A,m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-27 | 191 | 198 | 1A,m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-30-5p | 592 | 599 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-31 | 480 | 486 | 1A | hsa-miR-31 | AGGCAAGAUGCUGGCAUAGCUG |
| miR-329 | 820 | 826 | 1A | hsa-miR-329brain | AACACACCUGGUUAACCUCUUU |
| miR-330 | 117 | 124 | 1A,m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-338 | 61 | 67 | m8 | hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA |
| miR-365 | 302 | 309 | 1A,m8 | hsa-miR-365 | UAAUGCCCCUAAAAAUCCUUAU |
| miR-381 | 1052 | 1058 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-382 | 630 | 636 | m8 | hsa-miR-382brain | GAAGUUGUUCGUGGUGGAUUCG |
| miR-384 | 424 | 430 | m8 | hsa-miR-384 | AUUCCUAGAAAUUGUUCAUA |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 658 | 664 | 1A | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
| miR-96 | 482 | 488 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.