Gene Page: SMC3
Summary ?
| GeneID | 9126 |
| Symbol | SMC3 |
| Synonyms | BAM|BMH|CDLS3|CSPG6|HCAP|SMC3L1 |
| Description | structural maintenance of chromosomes 3 |
| Reference | MIM:606062|HGNC:HGNC:2468|Ensembl:ENSG00000108055|HPRD:05829|Vega:OTTHUMG00000019042 |
| Gene type | protein-coding |
| Map location | 10q25 |
| Pascal p-value | 0.049 |
| Sherlock p-value | 0.411 |
| Fetal beta | 2.154 |
| eGene | Myers' cis & trans |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.6061 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2083927 | chr3 | 66884108 | SMC3 | 9126 | 0.05 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
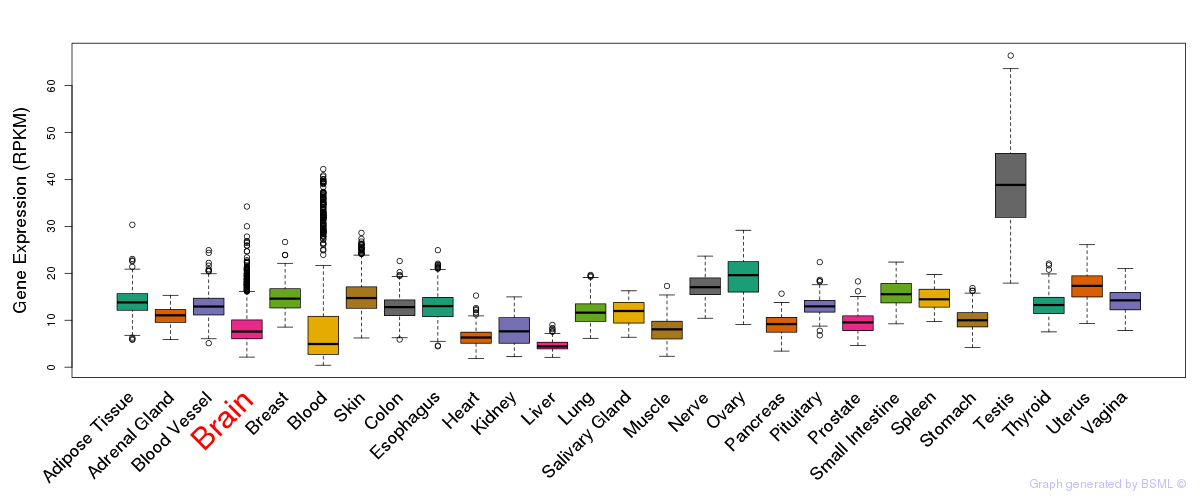
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LINGO1 | 0.93 | 0.94 |
| DMWD | 0.89 | 0.89 |
| MCOLN1 | 0.89 | 0.88 |
| FAM160A2 | 0.89 | 0.89 |
| CACNB3 | 0.88 | 0.89 |
| EPN1 | 0.88 | 0.89 |
| GSK3A | 0.88 | 0.89 |
| APBB1 | 0.87 | 0.89 |
| CLIP3 | 0.87 | 0.87 |
| DLGAP4 | 0.87 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.68 | -0.63 |
| AF347015.31 | -0.67 | -0.62 |
| AF347015.21 | -0.67 | -0.67 |
| AF347015.8 | -0.67 | -0.63 |
| AL139819.3 | -0.66 | -0.67 |
| AF347015.33 | -0.66 | -0.60 |
| MT-CYB | -0.66 | -0.61 |
| AF347015.27 | -0.65 | -0.63 |
| AF347015.26 | -0.64 | -0.59 |
| AP002478.3 | -0.64 | -0.65 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003777 | microtubule motor activity | NAS | 11590136 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016887 | ATPase activity | IEA | - | |
| GO:0046982 | protein heterodimerization activity | IPI | 11590136 | |
| GO:0045502 | dynein binding | IDA | 11590136 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006281 | DNA repair | IEA | - | |
| GO:0007165 | signal transduction | IDA | 12651860 | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0007062 | sister chromatid cohesion | NAS | 11590136 | |
| GO:0007067 | mitosis | TAS | 9506951 | |
| GO:0007126 | meiosis | IDA | 12498344 | |
| GO:0009294 | DNA mediated transformation | TAS | 10801778 | |
| GO:0007052 | mitotic spindle organization | IEP | 11590136 | |
| GO:0051301 | cell division | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000800 | lateral element | IEA | - | |
| GO:0000922 | spindle pole | IDA | 11590136 | |
| GO:0005604 | basement membrane | TAS | 9015313 | |
| GO:0005634 | nucleus | TAS | 9506951 | |
| GO:0005694 | chromosome | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 11590136 | |
| GO:0008278 | cohesin complex | NAS | 9506951 | |
| GO:0016363 | nuclear matrix | IDA | 11590136 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CASP4 | ICE(rel)II | ICEREL-II | ICH-2 | Mih1/TX | TX | caspase 4, apoptosis-related cysteine peptidase | Affinity Capture-MS | BioGRID | 17353931 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | Affinity Capture-MS | BioGRID | 17353931 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | Co-purification | BioGRID | 12198550 |
| KIFAP3 | FLJ22818 | KAP3 | SMAP | Smg-GDS | dJ190I16.1 | kinesin-associated protein 3 | - | HPRD,BioGRID | 9506951 |
| MAGED1 | DLXIN-1 | NRAGE | melanoma antigen family D, 1 | Affinity Capture-MS | BioGRID | 17353931 |
| MXD1 | MAD | MAD1 | MGC104659 | MAX dimerization protein 1 | Reconstituted Complex Two-hybrid | BioGRID | 9528857 |
| MXD3 | MAD3 | MGC2383 | bHLHc13 | MAX dimerization protein 3 | Reconstituted Complex Two-hybrid | BioGRID | 9528857 |
| MXD4 | MAD4 | MST149 | MSTP149 | bHLHc12 | MAX dimerization protein 4 | Reconstituted Complex Two-hybrid | BioGRID | 9528857 |
| MXI1 | MAD2 | MGC43220 | MXD2 | MXI | bHLHc11 | MAX interactor 1 | Reconstituted Complex Two-hybrid | BioGRID | 9528857 |
| NEK6 | SID6-1512 | NIMA (never in mitosis gene a)-related kinase 6 | Affinity Capture-MS | BioGRID | 17353931 |
| NUMA1 | NUMA | nuclear mitotic apparatus protein 1 | Affinity Capture-Western | BioGRID | 11590136 |
| REC8 | HR21spB | MGC950 | REC8L1 | Rec8p | REC8 homolog (yeast) | Affinity Capture-Western | BioGRID | 12759374 |
| SMARCA5 | ISWI | SNF2H | WCRF135 | hISWI | hSNF2H | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 | Co-purification | BioGRID | 12198550 |
| SMC1A | CDLS2 | DKFZp686L19178 | DXS423E | KIAA0178 | MGC138332 | SB1.8 | SMC1 | SMC1L1 | SMC1alpha | SMCB | structural maintenance of chromosomes 1A | Affinity Capture-Western | BioGRID | 9789013 |11590136 |11877376 |12759374 |
| SMC1A | CDLS2 | DKFZp686L19178 | DXS423E | KIAA0178 | MGC138332 | SB1.8 | SMC1 | SMC1L1 | SMC1alpha | SMCB | structural maintenance of chromosomes 1A | - | HPRD | 9789013 |11076961 |
| SMC1B | FLJ43748 | SMC1BETA | SMC1L2 | bK268H5 | bK268H5.5 | structural maintenance of chromosomes 1B | - | HPRD | 11564881 |12759374 |15125634 |
| STAG1 | DKFZp781D1416 | SA1 | stromal antigen 1 | Co-purification | BioGRID | 11076961 |
| SYCP3 | COR1 | MGC71888 | SCP3 | synaptonemal complex protein 3 | Affinity Capture-Western | BioGRID | 12759374 |
| TUBB | M40 | MGC117247 | MGC16435 | OK/SW-cl.56 | TUBB1 | TUBB5 | tubulin, beta | Affinity Capture-Western | BioGRID | 11590136 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| PID ATM PATHWAY | 34 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOSIS | 116 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME CHROMOSOME MAINTENANCE | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC PROMETAPHASE | 87 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOTIC SYNAPSIS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL DIVIDING UP | 57 | 33 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN DN | 27 | 20 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| PUJANA XPRSS INT NETWORK | 168 | 103 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA CENTERED NETWORK | 117 | 72 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF DN | 103 | 64 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES | 147 | 87 | All SZGR 2.0 genes in this pathway |
| OKUMURA INFLAMMATORY RESPONSE LPS | 183 | 115 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| BILD CTNNB1 ONCOGENIC SIGNATURE | 82 | 52 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR DN | 185 | 116 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| LI DCP2 BOUND MRNA | 89 | 57 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE EARLY LATE | 317 | 190 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS DN | 187 | 109 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| ABRAMSON INTERACT WITH AIRE | 45 | 33 | All SZGR 2.0 genes in this pathway |