Gene Page: CACNA2D2
Summary ?
| GeneID | 9254 |
| Symbol | CACNA2D2 |
| Synonyms | CACNA2D |
| Description | calcium voltage-gated channel auxiliary subunit alpha2delta 2 |
| Reference | MIM:607082|HGNC:HGNC:1400|Ensembl:ENSG00000007402|HPRD:06154|Vega:OTTHUMG00000156887 |
| Gene type | protein-coding |
| Map location | 3p21.3 |
| Pascal p-value | 0.07 |
| Sherlock p-value | 0.139 |
| Fetal beta | -0.979 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | CANABINOID DOPAMINE EXCITABILITY METABOTROPIC GLUTAMATE RECEPTOR SEROTONIN G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg21368467 | 3 | 50537375 | CACNA2D2 | 1.242E-4 | 0.309 | 0.03 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | CACNA2D2 | 9254 | 4.566E-5 | trans | ||
| rs7584986 | chr2 | 184111432 | CACNA2D2 | 9254 | 3.898E-4 | trans | ||
| rs17762315 | chr5 | 76807576 | CACNA2D2 | 9254 | 0.03 | trans | ||
| rs17795289 | chr7 | 43025774 | CACNA2D2 | 9254 | 0.13 | trans | ||
| rs16955618 | chr15 | 29937543 | CACNA2D2 | 9254 | 2.014E-8 | trans | ||
| rs159293 | chr17 | 32704562 | CACNA2D2 | 9254 | 0.11 | trans | ||
| rs212453 | 0 | CACNA2D2 | 9254 | 0.09 | trans | |||
| rs159261 | chr17 | 32718358 | CACNA2D2 | 9254 | 0.07 | trans | ||
| rs212489 | chr17 | 32730109 | CACNA2D2 | 9254 | 0.08 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
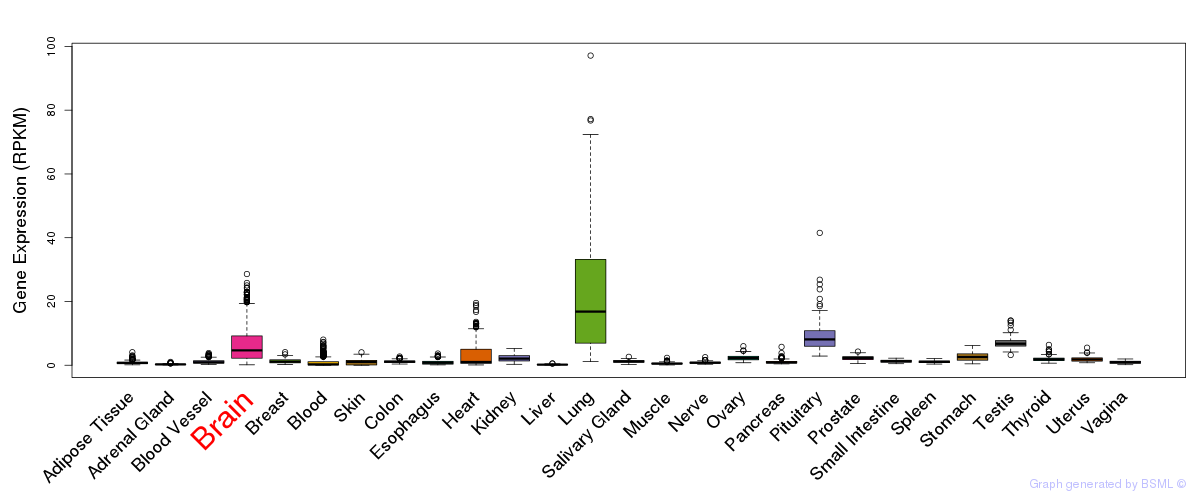
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ABCC8 | 0.55 | 0.55 |
| AL591845.1 | 0.52 | 0.51 |
| RGS11 | 0.51 | 0.49 |
| CES8 | 0.50 | 0.54 |
| IGFL4 | 0.49 | 0.50 |
| DNAH1 | 0.49 | 0.55 |
| CORO6 | 0.48 | 0.51 |
| CATSPER2 | 0.47 | 0.46 |
| DENND3 | 0.47 | 0.47 |
| SEC16B | 0.47 | 0.53 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RPSA | -0.31 | -0.30 |
| RPL35 | -0.30 | -0.40 |
| RPS6 | -0.30 | -0.34 |
| RPL15 | -0.30 | -0.28 |
| RPS3P3 | -0.30 | -0.33 |
| RPL18 | -0.29 | -0.37 |
| C9orf46 | -0.29 | -0.34 |
| RPS9 | -0.29 | -0.36 |
| RPS18 | -0.29 | -0.31 |
| RP11-568G20.1 | -0.29 | -0.28 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG CARDIAC MUSCLE CONTRACTION | 80 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG HYPERTROPHIC CARDIOMYOPATHY HCM | 85 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG ARRHYTHMOGENIC RIGHT VENTRICULAR CARDIOMYOPATHY ARVC | 76 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG DILATED CARDIOMYOPATHY | 92 | 68 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| HESSON TUMOR SUPPRESSOR CLUSTER 3P21 3 | 7 | 5 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| ROSS ACUTE MYELOID LEUKEMIA CBF | 82 | 57 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH AML1 ETO FUSION | 76 | 55 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA UP | 98 | 64 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER DN | 101 | 65 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 13 | 30 | 20 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| GREGORY SYNTHETIC LETHAL WITH IMATINIB | 145 | 83 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |