Gene Page: NRXN1
Summary ?
| GeneID | 9378 |
| Symbol | NRXN1 |
| Synonyms | Hs.22998|PTHSL2|SCZD17 |
| Description | neurexin 1 |
| Reference | MIM:600565|HGNC:HGNC:8008|Ensembl:ENSG00000179915|HPRD:11858|Vega:OTTHUMG00000129263 |
| Gene type | protein-coding |
| Map location | 2p16.3 |
| Sherlock p-value | 0.535 |
| TADA p-value | 0.025 |
| Fetal beta | 1.407 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | CELL ADHESION AND TRANSSYNAPTIC SIGNALING G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| DNM:Awadalla_2010 | Whole Exome Sequencing analysis | A survey of ~430 Mb of DNA from 401 synapse-expressed genes across all cases and 25 Mb of DNA in controls found 28 candidate DNMs. | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NRXN1 | chr2 | 50724807 | T | C | NM_001135659 NM_004801 | p.888E>G p.848E>G | missense missense | Schizophrenia | DNM:Fromer_2014 | ||
| NRXN1 | chr2 | 50149317 | - | ACGG | NRXN1:NM_138735:exon6:c.1093_1094insCCGT:p.T365fs NRXN1:NM_004801:exon22:c.4198_4199insCCGT:p.T1400fs NRXN1:NM_001135659:exon24:c.4408_4409insCCGT:p.T1470fs | G1402DfsX29 | frameshift insertion | Schizophrenia | DNM:Awadalla_2010 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg22324022 | 2 | 51259703 | NRXN1 | 2.157E-4 | 0.325 | 0.036 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs5909851 | chrX | 121070531 | NRXN1 | 9378 | 0.06 | trans |
Section II. Transcriptome annotation
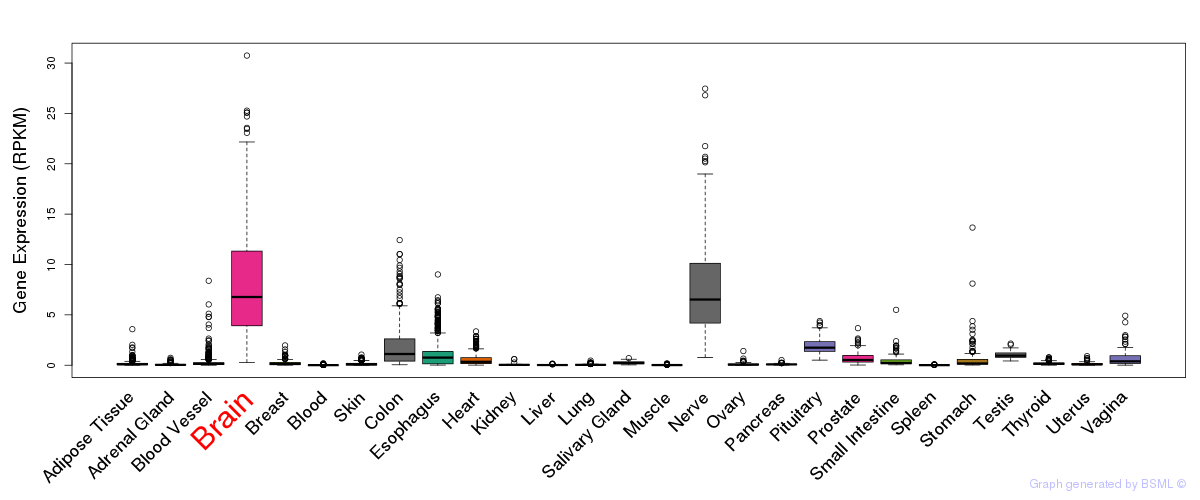
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PSMA1 | 0.90 | 0.87 |
| MRPS35 | 0.89 | 0.85 |
| OLA1 | 0.88 | 0.86 |
| ORC5L | 0.87 | 0.85 |
| STRAP | 0.87 | 0.84 |
| COPS3 | 0.86 | 0.82 |
| SKP1 | 0.86 | 0.84 |
| COPS4 | 0.86 | 0.85 |
| AZI2 | 0.85 | 0.83 |
| PMPCB | 0.85 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.2 | -0.69 | -0.59 |
| AF347015.26 | -0.68 | -0.60 |
| AF347015.8 | -0.67 | -0.57 |
| AF347015.15 | -0.65 | -0.56 |
| MT-CYB | -0.65 | -0.56 |
| MT-CO2 | -0.65 | -0.54 |
| AF347015.18 | -0.64 | -0.63 |
| AF347015.33 | -0.64 | -0.56 |
| AF347015.31 | -0.60 | -0.52 |
| AF347015.27 | -0.60 | -0.53 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | TAS | 1621094 | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 11152476 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007411 | axon guidance | TAS | axon (GO term level: 13) | 1621094 |
| GO:0007155 | cell adhesion | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 1621094 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APBA1 | D9S411E | MINT1 | X11 | X11A | X11ALPHA | amyloid beta (A4) precursor protein-binding, family A, member 1 | - | HPRD,BioGRID | 11036064 |
| APBA2 | D15S1518E | HsT16821 | LIN-10 | MGC99508 | MGC:14091 | MINT2 | X11L | amyloid beta (A4) precursor protein-binding, family A, member 2 | - | HPRD,BioGRID | 11036064 |
| CASK | CAGH39 | CMG | FLJ22219 | FLJ31914 | LIN2 | MICPCH | TNRC8 | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | - | HPRD,BioGRID | 11036064 |
| CASK | CAGH39 | CMG | FLJ22219 | FLJ31914 | LIN2 | MICPCH | TNRC8 | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | - | HPRD | 8786425 |
| MACF1 | ABP620 | ACF7 | FLJ45612 | FLJ46776 | KIAA0465 | KIAA1251 | MACF | OFC4 | microtubule-actin crosslinking factor 1 | - | HPRD | 12421765 |
| MLLT4 | AF-6 | AF6 | AFADIN | FLJ34371 | RP3-431P23.3 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4 | Two-hybrid | BioGRID | 9707552 |
| MYO16 | KIAA0865 | MYR8 | Myo16b | myosin XVI | - | HPRD | 12421765 |
| NLGN1 | KIAA1070 | MGC45115 | neuroligin 1 | - | HPRD,BioGRID | 8576240 |14522992 |
| NLGN2 | KIAA1366 | neuroligin 2 | - | HPRD,BioGRID | 8576240 |
| NLGN3 | ASPGX1 | AUTSX1 | HNL3 | KIAA1480 | neuroligin 3 | - | HPRD | 8576240 |9325340 |
| NLGN3 | ASPGX1 | AUTSX1 | HNL3 | KIAA1480 | neuroligin 3 | - | HPRD,BioGRID | 8576240 |
| NXPH1 | NPH1 | Nbla00697 | neurexophilin 1 | - | HPRD | 8699246 |
| NXPH1 | NPH1 | Nbla00697 | neurexophilin 1 | - | HPRD | 9856994 |
| NXPH2 | MGC125477 | MGC125478 | MGC125479 | MGC125480 | NPH2 | neurexophilin 2 | - | HPRD | 8699246 |
| NXPH3 | NPH3 | neurexophilin 3 | - | HPRD | 9856994 |
| PDZD2 | AIPC | KIAA0300 | PAPIN | PDZK3 | PIN1 | PDZ domain containing 2 | - | HPRD | 12421765 |
| RPH3A | KIAA0985 | rabphilin 3A homolog (mouse) | - | HPRD | 8901523 |
| SDCBP | MDA-9 | ST1 | SYCL | TACIP18 | syndecan binding protein (syntenin) | Neurexin I-alpha interacts with ST-1. This interaction was modelled on a demonstrated interaction between human neurexin I-alpha and rat ST-1. | BIND | 11152476 |
| SDCBP2 | FLJ12256 | SITAC18 | ST-2 | syndecan binding protein (syntenin) 2 | - | HPRD | 11152476 |
| SIPA1L1 | DKFZp686G1344 | E6TP1 | KIAA0440 | signal-induced proliferation-associated 1 like 1 | - | HPRD | 12421765 |
| SYT1 | DKFZp781D2042 | P65 | SVP65 | SYT | synaptotagmin I | - | HPRD | 8901523 |11243866 |
| SYT13 | KIAA1427 | synaptotagmin XIII | - | HPRD,BioGRID | 11171101 |
| SYT2 | SytII | synaptotagmin II | - | HPRD | 8901523 |
| SYT4 | HsT1192 | KIAA1342 | synaptotagmin IV | - | HPRD | 8901523 |
| SYT5 | - | synaptotagmin V | - | HPRD | 8901523 |
| SYT6 | - | synaptotagmin VI | - | HPRD | 8901523 |
| SYT7 | IPCA-7 | MGC150517 | PCANAP7 | SYT-VII | synaptotagmin VII | - | HPRD | 8901523 |
| SYT9 | FLJ45896 | synaptotagmin IX | - | HPRD | 8901523 |
| SYTL1 | FLJ14996 | JFC1 | SLP1 | synaptotagmin-like 1 | - | HPRD | 11243866 |
| SYTL2 | CHR11SYT | KIAA1597 | MGC102768 | SGA72M | SLP2 | synaptotagmin-like 2 | - | HPRD | 11243866 |
| SYTL3 | MGC105130 | MGC118883 | MGC118884 | MGC118885 | SLP3 | synaptotagmin-like 3 | - | HPRD | 11243866 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL UP | 146 | 99 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS B UP | 26 | 16 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| NIELSEN SCHWANNOMA UP | 18 | 16 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS UP | 139 | 93 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 1528 | 1534 | 1A | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-128 | 779 | 785 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-129-5p | 188 | 194 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC | ||||
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-141/200a | 230 | 236 | m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-181 | 1022 | 1029 | 1A,m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-196 | 377 | 383 | 1A | hsa-miR-196a | UAGGUAGUUUCAUGUUGUUGG |
| hsa-miR-196b | UAGGUAGUUUCCUGUUGUUGG | ||||
| miR-218 | 88 | 94 | m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-23 | 636 | 643 | 1A,m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-27 | 779 | 785 | m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-323 | 636 | 642 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-326 | 438 | 444 | 1A | hsa-miR-326 | CCUCUGGGCCCUUCCUCCAG |
| miR-328 | 205 | 211 | 1A | hsa-miR-328brain | CUGGCCCUCUCUGCCCUUCCGU |
| miR-33 | 817 | 823 | 1A | hsa-miR-33 | GUGCAUUGUAGUUGCAUUG |
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| miR-335 | 163 | 169 | m8 | hsa-miR-335brain | UCAAGAGCAAUAACGAAAAAUGU |
| miR-339 | 105 | 112 | 1A,m8 | hsa-miR-339 | UCCCUGUCCUCCAGGAGCUCA |
| miR-377 | 1111 | 1117 | 1A | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-381 | 22 | 29 | 1A,m8 | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU | ||||
| miR-410 | 747 | 753 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-455 | 90 | 96 | 1A | hsa-miR-455 | UAUGUGCCUUUGGACUACAUCG |
| miR-485-3p | 1018 | 1025 | 1A,m8 | hsa-miR-485-3p | GUCAUACACGGCUCUCCUCUCU |
| hsa-miR-485-3p | GUCAUACACGGCUCUCCUCUCU | ||||
| miR-96 | 1326 | 1332 | m8 | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.