Gene Page: EIF2AK3
Summary ?
| GeneID | 9451 |
| Symbol | EIF2AK3 |
| Synonyms | PEK|PERK|WRS |
| Description | eukaryotic translation initiation factor 2 alpha kinase 3 |
| Reference | MIM:604032|HGNC:HGNC:3255|Ensembl:ENSG00000172071|HPRD:04942|Vega:OTTHUMG00000155046 |
| Gene type | protein-coding |
| Map location | 2p12 |
| Pascal p-value | 0.054 |
| Fetal beta | 0.719 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0004 | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg05085631 | 2 | 88927217 | EIF2AK3 | 1.47E-9 | -0.013 | 1.39E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
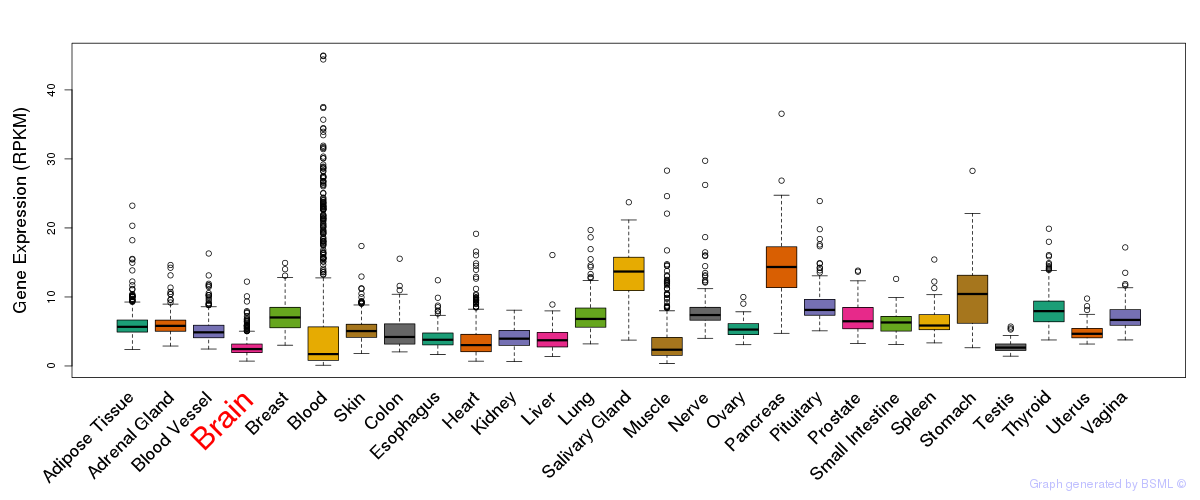
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity | IDA | 10026192 |10677345 |11907036 | |
| GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity | IMP | 12086964 | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0042802 | identical protein binding | IPI | 11907036 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0031642 | negative regulation of myelination | IEA | axon (GO term level: 15) | - |
| GO:0001501 | skeletal system development | IEA | - | |
| GO:0002063 | chondrocyte development | IEA | - | |
| GO:0006986 | response to unfolded protein | IEA | - | |
| GO:0048009 | insulin-like growth factor receptor signaling pathway | IEA | - | |
| GO:0007029 | endoplasmic reticulum organization | IEA | - | |
| GO:0006983 | ER overload response | IDA | 10677345 |11907036 | |
| GO:0006926 | virus-infected cell apoptosis | IEA | - | |
| GO:0006919 | caspase activation | IEA | - | |
| GO:0019722 | calcium-mediated signaling | IEA | - | |
| GO:0030073 | insulin secretion | IEA | - | |
| GO:0031018 | endocrine pancreas development | IMP | 12086964 | |
| GO:0032092 | positive regulation of protein binding | IEA | - | |
| GO:0030968 | endoplasmic reticulum unfolded protein response | IDA | 10677345 |11907036 | |
| GO:0032057 | negative regulation of translational initiation in response to stress | TAS | 10677345 |12086964 | |
| GO:0051260 | protein homooligomerization | IMP | 11907036 | |
| GO:0030282 | bone mineralization | IEA | - | |
| GO:0017148 | negative regulation of translation | TAS | 12086964 | |
| GO:0046777 | protein amino acid autophosphorylation | IDA | 10026192 |11907036 | |
| GO:0046777 | protein amino acid autophosphorylation | IMP | 12086964 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005789 | endoplasmic reticulum membrane | NAS | 11907036 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005783 | endoplasmic reticulum | IC | 11907036 | |
| GO:0005783 | endoplasmic reticulum | TAS | 12086964 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EIF2 PATHWAY | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME DIABETES PATHWAYS | 133 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME PERK REGULATED GENE EXPRESSION | 29 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME UNFOLDED PROTEIN RESPONSE | 80 | 51 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| MAYBURD RESPONSE TO L663536 UP | 29 | 18 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM2 | 153 | 102 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX UP | 83 | 66 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| PODAR RESPONSE TO ADAPHOSTIN UP | 147 | 98 | All SZGR 2.0 genes in this pathway |
| CUI GLUCOSE DEPRIVATION | 60 | 44 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| HOFFMANN IMMATURE TO MATURE B LYMPHOCYTE UP | 43 | 34 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 UP | 87 | 52 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 3 TRANSIENTLY INDUCED BY EGF | 222 | 159 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-130/301 | 478 | 484 | 1A | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.