Gene Page: NRG2
Summary ?
| GeneID | 9542 |
| Symbol | NRG2 |
| Synonyms | DON1|HRG2|NTAK |
| Description | neuregulin 2 |
| Reference | MIM:603818|HGNC:HGNC:7998|Ensembl:ENSG00000158458|HPRD:04821| |
| Gene type | protein-coding |
| Map location | 5q31.2 |
| Pascal p-value | 0.032 |
| Fetal beta | 0.448 |
| DMG | 2 (# studies) |
| Support | NEUROTROPHIN SIGNALING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15992535 | 5 | 139228150 | NRG2 | 5.466E-4 | 1.142 | 0.049 | DMG:Wockner_2014 |
| cg27152280 | 5 | 139423077 | NRG2 | 3.81E-9 | -0.008 | 2.4E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
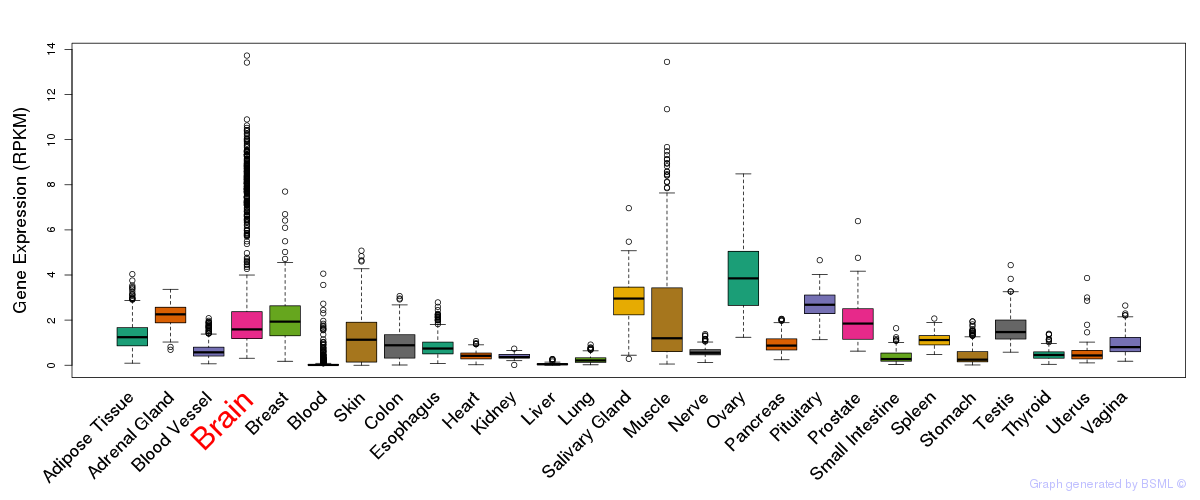
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| UNK | 0.90 | 0.93 |
| MEN1 | 0.89 | 0.92 |
| U2AF2 | 0.88 | 0.91 |
| BANP | 0.88 | 0.90 |
| NUP62 | 0.88 | 0.93 |
| CCDC97 | 0.88 | 0.92 |
| MTA1 | 0.88 | 0.90 |
| DVL3 | 0.88 | 0.91 |
| MAP3K7IP1 | 0.88 | 0.91 |
| TTYH3 | 0.87 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.72 | -0.87 |
| MT-CO2 | -0.71 | -0.87 |
| AF347015.27 | -0.70 | -0.86 |
| AF347015.33 | -0.69 | -0.83 |
| C5orf53 | -0.69 | -0.76 |
| MT-CYB | -0.69 | -0.84 |
| S100B | -0.68 | -0.79 |
| FXYD1 | -0.67 | -0.81 |
| AF347015.8 | -0.67 | -0.85 |
| AF347015.21 | -0.65 | -0.88 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008083 | growth factor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007165 | signal transduction | TAS | 9168114 |9168115 | |
| GO:0009790 | embryonic development | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ERBB SIGNALING PATHWAY | 87 | 71 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AGR PATHWAY | 36 | 31 | All SZGR 2.0 genes in this pathway |
| PID ERBB4 PATHWAY | 38 | 32 | All SZGR 2.0 genes in this pathway |
| PID ERBB2 ERBB3 PATHWAY | 44 | 35 | All SZGR 2.0 genes in this pathway |
| PID ERBB NETWORK PATHWAY | 15 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | 22 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB4 SIGNALING | 38 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | 20 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB2 SIGNALING | 44 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR SIGNALING BY ERBB4 | 38 | 30 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE UP | 283 | 177 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| VISALA AGING LYMPHOCYTE DN | 19 | 10 | All SZGR 2.0 genes in this pathway |
| KUROKAWA LIVER CANCER EARLY RECURRENCE UP | 12 | 9 | All SZGR 2.0 genes in this pathway |
| GREGORY SYNTHETIC LETHAL WITH IMATINIB | 145 | 83 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-129-5p | 155 | 161 | m8 | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-450 | 155 | 161 | 1A | hsa-miR-450 | UUUUUGCGAUGUGUUCCUAAUA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.