Gene Page: TRAF4
Summary ?
| GeneID | 9618 |
| Symbol | TRAF4 |
| Synonyms | CART1|MLN62|RNF83 |
| Description | TNF receptor associated factor 4 |
| Reference | MIM:602464|HGNC:HGNC:12034|Ensembl:ENSG00000076604|HPRD:03915|Vega:OTTHUMG00000132677 |
| Gene type | protein-coding |
| Map location | 17q11-q12 |
| Pascal p-value | 0.274 |
| Sherlock p-value | 0.062 |
| Fetal beta | 1.447 |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2070265 | chr17 | 27075422 | TRAF4 | 9618 | 0.16 | cis | ||
| rs4794836 | chr17 | 27082709 | TRAF4 | 9618 | 0.08 | cis | ||
| rs4795457 | chr17 | 27087928 | TRAF4 | 9618 | 0.14 | cis | ||
| rs11658884 | chr17 | 27132741 | TRAF4 | 9618 | 0.12 | cis | ||
| rs2242345 | chr17 | 27185826 | TRAF4 | 9618 | 0.08 | cis | ||
| rs2277664 | chr17 | 27286661 | TRAF4 | 9618 | 0.18 | cis |
Section II. Transcriptome annotation
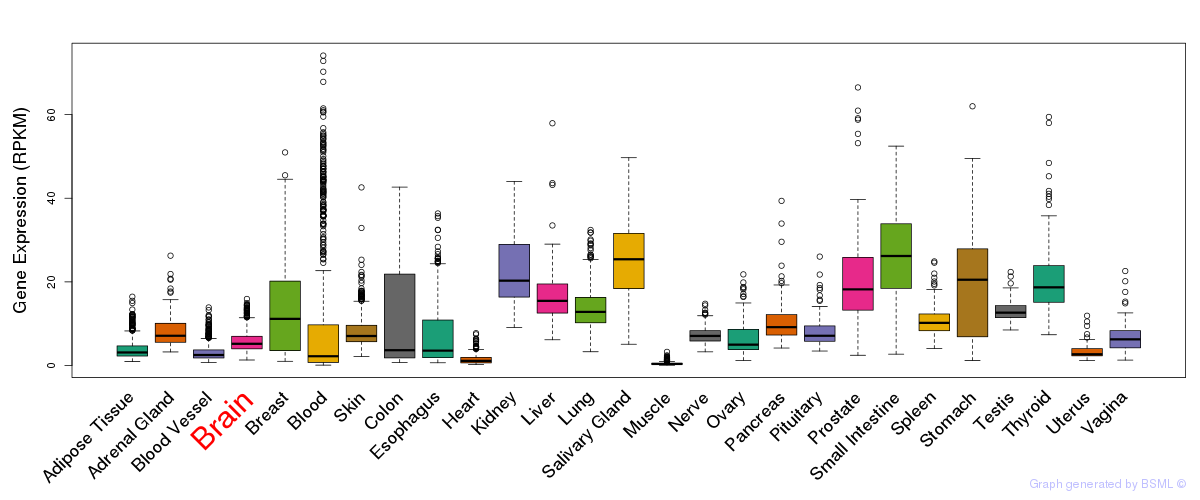
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PPP1R3F | 0.92 | 0.94 |
| ABLIM2 | 0.92 | 0.94 |
| ZFYVE28 | 0.91 | 0.88 |
| AC105206.1 | 0.91 | 0.91 |
| HTR5A | 0.89 | 0.91 |
| PRRT3 | 0.89 | 0.94 |
| NPTN | 0.89 | 0.90 |
| C2orf55 | 0.89 | 0.79 |
| MAST3 | 0.89 | 0.92 |
| SYNPO | 0.89 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C9orf46 | -0.46 | -0.47 |
| GTF3C6 | -0.46 | -0.43 |
| RBMX2 | -0.44 | -0.47 |
| EXOSC8 | -0.44 | -0.37 |
| BCL7C | -0.44 | -0.48 |
| RPL23A | -0.44 | -0.47 |
| RPS8 | -0.43 | -0.44 |
| RPS20 | -0.42 | -0.49 |
| RPS6 | -0.42 | -0.44 |
| DYNLT1 | -0.42 | -0.48 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTN1 | FLJ40884 | actinin, alpha 1 | TRAF4 interacts with alpha-actinin. | BIND | 12023963 |
| BCKDK | - | branched chain ketoacid dehydrogenase kinase | TRAF4 interacts with BCKDK. | BIND | 12023963 |
| CCHCR1 | C6orf18 | HCR | MGC126371 | MGC126372 | SBP | coiled-coil alpha-helical rod protein 1 | TRAF4 interacts with HCR. | BIND | 12023963 |
| LTBR | CD18 | D12S370 | LT-BETA-R | TNF-R-III | TNFCR | TNFR-RP | TNFR2-RP | TNFRSF3 | lymphotoxin beta receptor (TNFR superfamily, member 3) | - | HPRD,BioGRID | 9626059 |
| MAGED1 | DLXIN-1 | NRAGE | melanoma antigen family D, 1 | TRAF4 interacts with NRAGE. | BIND | 12023963 |
| ME3 | FLJ34862 | malic enzyme 3, NADP(+)-dependent, mitochondrial | TRAF4 interacts with NADP-ME. | BIND | 12023963 |
| NCF1 | FLJ79451 | NCF1A | NOXO2 | SH3PXD1A | p47phox | neutrophil cytosolic factor 1 | Affinity Capture-Western | BioGRID | 12023963 |
| NCF1C | SH3PXD1C | neutrophil cytosolic factor 1C pseudogene | TRAF4 interacts with p47phox. | BIND | 12023963 |
| NCF1C | SH3PXD1C | neutrophil cytosolic factor 1C pseudogene | - | HPRD | 12023963 |
| NGFR | CD271 | Gp80-LNGFR | TNFRSF16 | p75(NTR) | p75NTR | nerve growth factor receptor (TNFR superfamily, member 16) | - | HPRD,BioGRID | 10514511 |
| PLSCR1 | MMTRA1B | phospholipid scramblase 1 | TRAF4 interacts with PLSCR1. | BIND | 12023963 |
| PXDN | D2S448 | D2S448E | KIAA0230 | MG50 | PRG2 | PXN | VPO | peroxidasin homolog (Drosophila) | TRAF4 interacts with MAGE. | BIND | 12023963 |
| RPS6KB1 | PS6K | S6K | S6K1 | STK14A | p70(S6K)-alpha | p70-S6K | p70-alpha | ribosomal protein S6 kinase, 70kDa, polypeptide 1 | - | HPRD,BioGRID | 12801526 |
| SIGLEC7 | AIRM1 | CD328 | CDw328 | D-siglec | QA79 | SIGLEC-7 | p75 | p75/AIRM1 | sialic acid binding Ig-like lectin 7 | - | HPRD | 10514511 |
| SORBS2 | ARGBP2 | FLJ93447 | KIAA0777 | PRO0618 | sorbin and SH3 domain containing 2 | TRAF4 interacts with ArgBP2. | BIND | 12023963 |
| TARBP2 | LOQS | TRBP | TRBP1 | TRBP2 | TAR (HIV-1) RNA binding protein 2 | TRAF4 interacts with TARBP2. | BIND | 12023963 |
| TBC1D8 | AD3 | HBLP1 | VRP | TBC1 domain family, member 8 (with GRAM domain) | TRAF4 interacts with VRP. | BIND | 12023963 |
| TGFB1I1 | ARA55 | HIC-5 | HIC5 | TSC-5 | transforming growth factor beta 1 induced transcript 1 | TRAF4 interacts with Hic-5. | BIND | 12023963 |
| TRIM37 | KIAA0898 | MUL | POB1 | TEF3 | tripartite motif-containing 37 | - | HPRD,BioGRID | 11279055 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | TRAF4 interacts with UBC9. | BIND | 12023963 |
| USP7 | HAUSP | TEF1 | ubiquitin specific peptidase 7 (herpes virus-associated) | - | HPRD,BioGRID | 11279055 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| PID TAP63 PATHWAY | 54 | 40 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP UP | 265 | 158 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| LAU APOPTOSIS CDKN2A UP | 55 | 40 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 UP | 150 | 93 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL DN | 186 | 107 | All SZGR 2.0 genes in this pathway |
| LUCAS HNF4A TARGETS UP | 58 | 36 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER | 178 | 108 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE EARLY | 66 | 48 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA | 164 | 118 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 7 | 27 | 22 | All SZGR 2.0 genes in this pathway |
| MORI EMU MYC LYMPHOMA BY ONSET TIME UP | 110 | 69 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q11 Q21 AMPLICON | 133 | 78 | All SZGR 2.0 genes in this pathway |
| SCHURINGA STAT5A TARGETS UP | 21 | 13 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR DN | 63 | 41 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| RHODES CANCER META SIGNATURE | 64 | 47 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC DN | 123 | 86 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C0 | 107 | 72 | All SZGR 2.0 genes in this pathway |
| JACKSON DNMT1 TARGETS UP | 77 | 57 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER ERBB2 UP | 147 | 83 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 12HR DN | 33 | 25 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 24HR DN | 43 | 32 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 32HR DN | 39 | 28 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 48HR DN | 40 | 29 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE UP | 242 | 159 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE MIDDLE | 98 | 56 | All SZGR 2.0 genes in this pathway |
| CHANGOLKAR H2AFY TARGETS UP | 48 | 28 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 COMPLETE | 227 | 151 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR UP | 166 | 97 | All SZGR 2.0 genes in this pathway |