Gene Page: CD79A
Summary ?
| GeneID | 973 |
| Symbol | CD79A |
| Synonyms | IGA|MB-1 |
| Description | CD79a molecule |
| Reference | MIM:112205|HGNC:HGNC:1698|Ensembl:ENSG00000105369|HPRD:00203|Vega:OTTHUMG00000182678 |
| Gene type | protein-coding |
| Map location | 19q13.2 |
| Pascal p-value | 0.301 |
| Fetal beta | -0.294 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg11719283 | 19 | 42574717 | CD79A | 8.41E-4 | 10.837 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10994209 | chr10 | 61877705 | CD79A | 973 | 0.11 | trans |
Section II. Transcriptome annotation
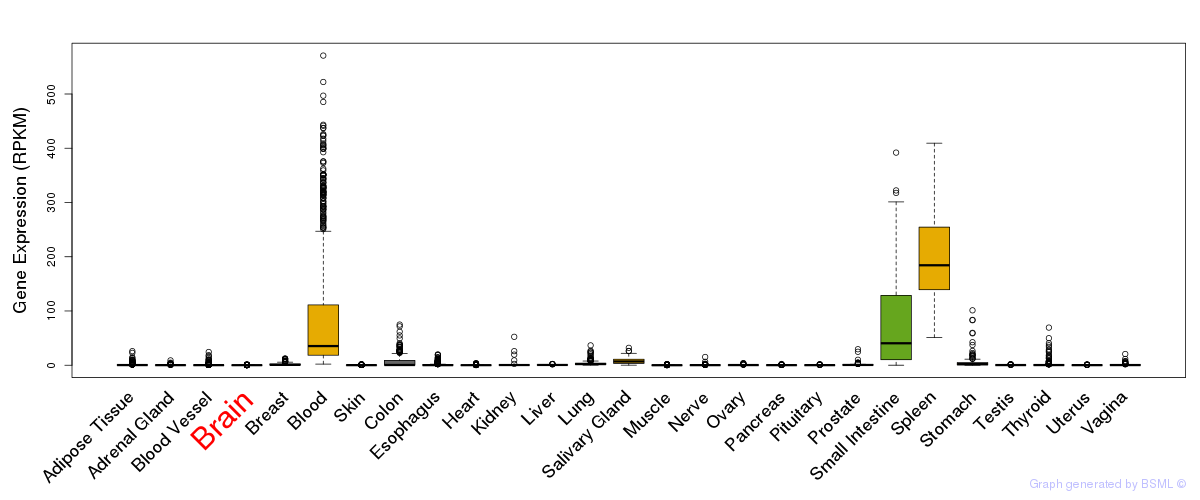
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| IFITM2 | 0.57 | 0.50 |
| MT2A | 0.56 | 0.44 |
| MT1F | 0.54 | 0.41 |
| CEBPD | 0.53 | 0.42 |
| MT1X | 0.52 | 0.43 |
| IFITM3 | 0.52 | 0.48 |
| RARRES3 | 0.52 | 0.39 |
| TREX1 | 0.52 | 0.44 |
| SRGN | 0.52 | 0.43 |
| MT1M | 0.52 | 0.41 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CCDC49 | -0.37 | -0.38 |
| ZNF638 | -0.36 | -0.36 |
| ARID4A | -0.36 | -0.38 |
| ING2 | -0.35 | -0.39 |
| RTF1 | -0.35 | -0.34 |
| USP7 | -0.35 | -0.34 |
| GIGYF2 | -0.35 | -0.33 |
| ZNF417 | -0.35 | -0.34 |
| STX3 | -0.35 | -0.35 |
| HIVEP3 | -0.34 | -0.37 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004888 | transmembrane receptor activity | IEA | - | |
| GO:0005515 | protein binding | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007166 | cell surface receptor linked signal transduction | IEA | - | |
| GO:0006955 | immune response | IEA | - | |
| GO:0042100 | B cell proliferation | ISS | - | |
| GO:0030183 | B cell differentiation | ISS | - | |
| GO:0050853 | B cell receptor signaling pathway | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0019815 | B cell receptor complex | ISS | Synap (GO term level: 8) | - |
| GO:0005771 | multivesicular body | ISS | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0009897 | external side of plasma membrane | ISS | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0045121 | membrane raft | ISS | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AMBP | EDC1 | HCP | HI30 | IATIL | ITI | ITIL | ITILC | UTI | alpha-1-microglobulin/bikunin precursor | Affinity Capture-Western Co-purification | BioGRID | 6196366 |9183005 |
| BLK | MGC10442 | B lymphoid tyrosine kinase | - | HPRD,BioGRID | 7592958 |
| BLNK | BASH | BLNK-S | LY57 | MGC111051 | SLP-65 | SLP65 | B-cell linker | - | HPRD,BioGRID | 11449366 |11909947 |
| CD19 | B4 | MGC12802 | CD19 molecule | - | HPRD,BioGRID | 9120258 |
| CD79A | IGA | MB-1 | CD79a molecule, immunoglobulin-associated alpha | - | HPRD | 1881434 |
| CD79B | B29 | IGB | CD79b molecule, immunoglobulin-associated beta | Affinity Capture-Western | BioGRID | 12886015 |
| CD79B | B29 | IGB | CD79b molecule, immunoglobulin-associated beta | - | HPRD | 1591006 |8656670 |
| FANCA | FA | FA-H | FA1 | FAA | FACA | FAH | FANCH | MGC75158 | Fanconi anemia, complementation group A | Two-hybrid | BioGRID | 14499622 |
| FCAR | CD89 | Fc fragment of IgA, receptor for | - | HPRD | 8666916 |10438530 |
| HCLS1 | CTTNL | HS1 | hematopoietic cell-specific Lyn substrate 1 | - | HPRD,BioGRID | 7927516 |
| IGHM | DKFZp686I15196 | DKFZp686I15212 | FLJ00385 | MGC104996 | MGC52291 | MU | VH | immunoglobulin heavy constant mu | - | HPRD | 9561918 |
| IGJ | IGCJ | JCH | immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides | - | HPRD | 1292512 |8683109 |
| PIGR | FLJ22667 | MGC125361 | MGC125362 | polymeric immunoglobulin receptor | - | HPRD | 1940346 |7989333 |10540352 |12186846 |
| PTPRC | B220 | CD45 | CD45R | GP180 | LCA | LY5 | T200 | protein tyrosine phosphatase, receptor type, C | - | HPRD,BioGRID | 7516335 |
| SYK | DKFZp313N1010 | FLJ25043 | FLJ37489 | spleen tyrosine kinase | - | HPRD,BioGRID | 1569106|7500027 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG B CELL RECEPTOR SIGNALING PATHWAY | 75 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PRIMARY IMMUNODEFICIENCY | 35 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BCR PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CTCF PATHWAY | 23 | 18 | All SZGR 2.0 genes in this pathway |
| PID BCR 5PATHWAY | 65 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | 29 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA DN | 52 | 30 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA DN | 104 | 59 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| KLEIN TARGETS OF BCR ABL1 FUSION | 45 | 34 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE DN | 165 | 104 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 9 | 19 | 13 | All SZGR 2.0 genes in this pathway |
| GOLUB ALL VS AML UP | 24 | 20 | All SZGR 2.0 genes in this pathway |
| KLEIN PRIMARY EFFUSION LYMPHOMA DN | 58 | 42 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| ZHANG TARGETS OF EWSR1 FLI1 FUSION | 88 | 68 | All SZGR 2.0 genes in this pathway |
| ASTIER INTEGRIN SIGNALING | 59 | 44 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD2 UP | 45 | 32 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| KRASNOSELSKAYA ILF3 TARGETS UP | 38 | 24 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES DN | 245 | 144 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES UP | 253 | 147 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER UP | 288 | 168 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS LCP WITH H3K4ME3 | 174 | 100 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |