Gene Page: BCL2L11
Summary ?
| GeneID | 10018 |
| Symbol | BCL2L11 |
| Synonyms | BAM|BIM|BOD |
| Description | BCL2 like 11 |
| Reference | MIM:603827|HGNC:HGNC:994|Ensembl:ENSG00000153094|HPRD:04828|Vega:OTTHUMG00000131256 |
| Gene type | protein-coding |
| Map location | 2q13 |
| Pascal p-value | 0.029 |
| Fetal beta | 0.668 |
| eGene | Cerebellar Hemisphere Cerebellum |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0004 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00755 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
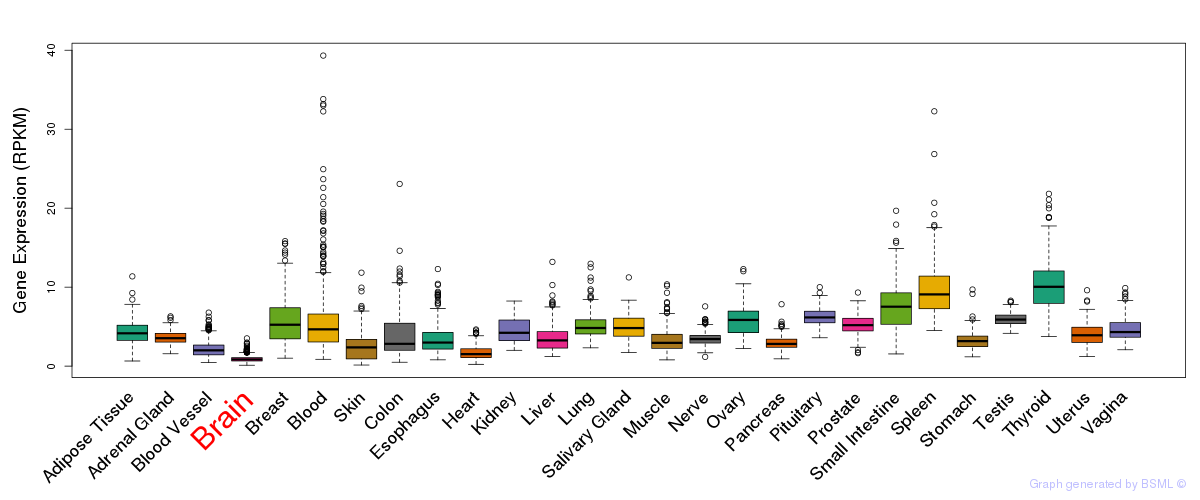
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| IQGAP1 | 0.81 | 0.76 |
| WWC3 | 0.81 | 0.81 |
| C1orf198 | 0.81 | 0.83 |
| NDE1 | 0.80 | 0.75 |
| WDFY2 | 0.80 | 0.79 |
| PHLPP1 | 0.79 | 0.78 |
| FLNB | 0.79 | 0.83 |
| SALL1 | 0.79 | 0.80 |
| CTNNA1 | 0.79 | 0.76 |
| FUBP3 | 0.78 | 0.76 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TTC9B | -0.43 | -0.46 |
| SYCP3 | -0.42 | -0.39 |
| ST20 | -0.39 | -0.43 |
| AC044839.2 | -0.39 | -0.32 |
| USMG5 | -0.39 | -0.45 |
| UQCR | -0.39 | -0.41 |
| UQCRQ | -0.39 | -0.41 |
| ZFAND2A | -0.39 | -0.41 |
| CARD16 | -0.38 | -0.38 |
| RERGL | -0.38 | -0.36 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 10198631 |15694340 |16282323 |17692808 | |
| GO:0008017 | microtubule binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007160 | cell-matrix adhesion | IEA | - | |
| GO:0006917 | induction of apoptosis | TAS | 10381623 | |
| GO:0008633 | activation of pro-apoptotic gene products | EXP | 14764673 | |
| GO:0030879 | mammary gland development | IEA | - | |
| GO:0035148 | lumen formation | IEA | - | |
| GO:0046620 | regulation of organ growth | IEA | - | |
| GO:0048563 | post-embryonic organ morphogenesis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 12591950 | |
| GO:0005624 | membrane fraction | TAS | 9430630 | |
| GO:0005741 | mitochondrial outer membrane | EXP | 10198631 | |
| GO:0005886 | plasma membrane | EXP | 12591950 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SA PROGRAMMED CELL DEATH | 12 | 9 | All SZGR 2.0 genes in this pathway |
| PID P73PATHWAY | 79 | 59 | All SZGR 2.0 genes in this pathway |
| PID FOXO PATHWAY | 49 | 43 | All SZGR 2.0 genes in this pathway |
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY | 70 | 60 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME NRAGE SIGNALS DEATH THROUGH JNK | 43 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | 60 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | 81 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF BH3 ONLY PROTEINS | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | 30 | 21 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| PIEPOLI LGI1 TARGETS UP | 15 | 11 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D UP | 175 | 108 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL DN | 226 | 132 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| LAU APOPTOSIS CDKN2A UP | 55 | 40 | All SZGR 2.0 genes in this pathway |
| CHEBOTAEV GR TARGETS UP | 77 | 62 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| RAMJAUN APOPTOSIS BY TGFB1 VIA SMAD4 UP | 7 | 6 | All SZGR 2.0 genes in this pathway |
| MARKS HDAC TARGETS UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 AND TP63 TARGETS | 205 | 145 | All SZGR 2.0 genes in this pathway |
| GALLUZZI PERMEABILIZE MITOCHONDRIA | 43 | 31 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 4 | 61 | 34 | All SZGR 2.0 genes in this pathway |
| YAN ESCAPE FROM ANOIKIS | 24 | 19 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| BROCKE APOPTOSIS REVERSED BY IL6 | 144 | 98 | All SZGR 2.0 genes in this pathway |
| OKUMURA INFLAMMATORY RESPONSE LPS | 183 | 115 | All SZGR 2.0 genes in this pathway |
| GALINDO IMMUNE RESPONSE TO ENTEROTOXIN | 85 | 67 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR UP | 101 | 69 | All SZGR 2.0 genes in this pathway |
| BILD E2F3 ONCOGENIC SIGNATURE | 246 | 153 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T7 | 98 | 63 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE DN | 61 | 39 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION UP | 180 | 114 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY DN | 58 | 39 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| LEE DIFFERENTIATING T LYMPHOCYTE | 200 | 115 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER UP | 288 | 168 | All SZGR 2.0 genes in this pathway |
| FONTAINE FOLLICULAR THYROID ADENOMA UP | 75 | 43 | All SZGR 2.0 genes in this pathway |
| FONTAINE THYROID TUMOR UNCERTAIN MALIGNANCY UP | 36 | 23 | All SZGR 2.0 genes in this pathway |
| PYEON CANCER HEAD AND NECK VS CERVICAL UP | 193 | 95 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 1 | 68 | 50 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR UP | 61 | 44 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA | 140 | 96 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS DN | 27 | 24 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
| VANOEVELEN MYOGENESIS SIN3A TARGETS | 220 | 133 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-10 | 4111 | 4118 | 1A,m8 | hsa-miR-10a | UACCCUGUAGAUCCGAAUUUGUG |
| hsa-miR-10b | UACCCUGUAGAACCGAAUUUGU | ||||
| miR-101 | 2255 | 2262 | 1A,m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-124.1 | 465 | 471 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 465 | 471 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA | ||||
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-130/301 | 4093 | 4099 | m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-144 | 2256 | 2262 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-148/152 | 4094 | 4101 | 1A,m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| hsa-miR-148a | UCAGUGCACUACAGAACUUUGU | ||||
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-149 | 444 | 450 | m8 | hsa-miR-149brain | UCUGGCUCCGUGUCUUCACUCC |
| miR-17-5p/20/93.mr/106/519.d | 1999 | 2006 | 1A,m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-181 | 208 | 215 | 1A,m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-19 | 4092 | 4098 | m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-214 | 3734 | 3741 | 1A,m8 | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-221/222 | 4052 | 4059 | 1A,m8 | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| miR-24 | 2678 | 2685 | 1A,m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG | ||||
| miR-25/32/92/363/367 | 4134 | 4141 | 1A,m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA | ||||
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-30-5p | 288 | 295 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-339 | 4110 | 4116 | m8 | hsa-miR-339 | UCCCUGUCCUCCAGGAGCUCA |
| miR-376 | 2184 | 2191 | 1A,m8 | hsa-miR-376a | AUCAUAGAGGAAAAUCCACGU |
| hsa-miR-376b | AUCAUAGAGGAAAAUCCAUGUU | ||||
| miR-381 | 2859 | 2865 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-496 | 211 | 217 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-9 | 1927 | 1934 | 1A,m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 1998 | 2004 | m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.