Gene Page: RANBP9
Summary ?
| GeneID | 10048 |
| Symbol | RANBP9 |
| Synonyms | BPM-L|BPM90|RANBPM|RanBP7 |
| Description | RAN binding protein 9 |
| Reference | MIM:603854|HGNC:HGNC:13727|Ensembl:ENSG00000010017|HPRD:04835|Vega:OTTHUMG00000015642 |
| Gene type | protein-coding |
| Map location | 6p23 |
| Pascal p-value | 0.012 |
| Sherlock p-value | 4.646E-5 |
| Fetal beta | -0.348 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0159 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0833 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
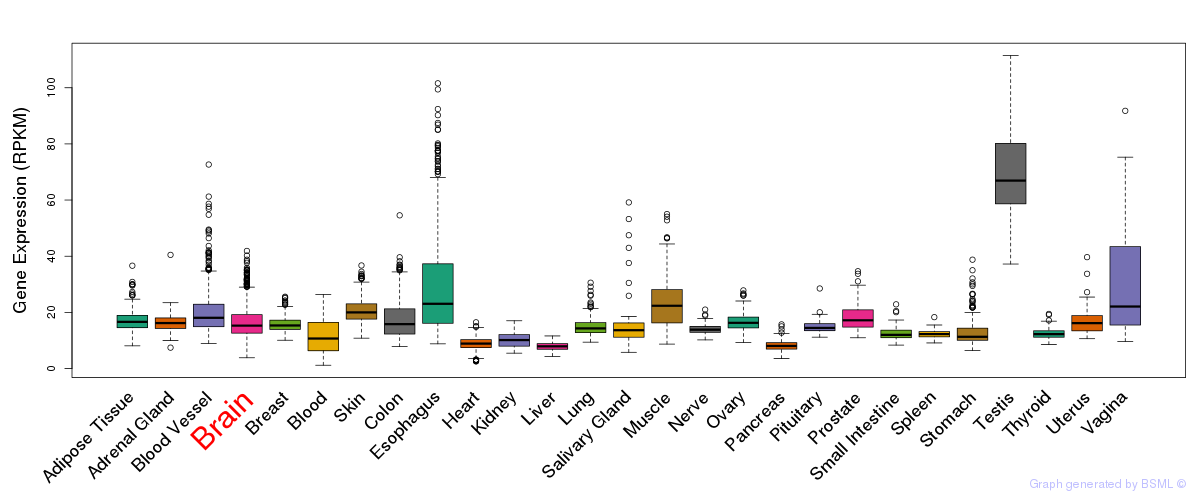
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MAP3K1 | 0.78 | 0.62 |
| AP001979.1 | 0.77 | 0.66 |
| TSPAN9 | 0.77 | 0.64 |
| SPATA13 | 0.77 | 0.67 |
| ZNF503 | 0.76 | 0.64 |
| TRERF1 | 0.76 | 0.77 |
| FOXP4 | 0.75 | 0.66 |
| RARA | 0.75 | 0.67 |
| MEIS1 | 0.75 | 0.63 |
| AC010997.1 | 0.75 | 0.59 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.45 | -0.47 |
| C1orf54 | -0.42 | -0.48 |
| SYCP3 | -0.41 | -0.49 |
| AF347015.31 | -0.40 | -0.41 |
| MT-CO2 | -0.40 | -0.40 |
| VAMP5 | -0.39 | -0.40 |
| GNG11 | -0.38 | -0.42 |
| HIGD1B | -0.37 | -0.36 |
| CLEC2B | -0.37 | -0.42 |
| IL32 | -0.37 | -0.45 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 12220523 |12421467 |14500717 | |
| GO:0008536 | Ran GTPase binding | TAS | 9817760 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006461 | protein complex assembly | TAS | 9817760 | |
| GO:0007020 | microtubule nucleation | NAS | 12220523 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005875 | microtubule associated complex | TAS | 9817760 | |
| GO:0005634 | nucleus | IDA | 12220523 | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD,BioGRID | 12361945 |
| AXL | JTK11 | UFO | AXL receptor tyrosine kinase | Axl interacts with RanBPM. | BIND | 12470648 |
| BACE1 | ASP2 | BACE | FLJ90568 | HSPC104 | KIAA1149 | beta-site APP-cleaving enzyme 1 | BACE1 interacts with RANBP9 (RANBPM). This interaction was modelled on a demonstrated interaction between mouse BACE1 and human RANBPM. | BIND | 15606899 |
| CALB1 | CALB | calbindin 1, 28kDa | - | HPRD,BioGRID | 12684061 |
| DISC1 | C1orf136 | FLJ13381 | FLJ21640 | FLJ25311 | FLJ41105 | KIAA0457 | SCZD9 | disrupted in schizophrenia 1 | - | HPRD,BioGRID | 12812986 |
| DISC1 | C1orf136 | FLJ13381 | FLJ21640 | FLJ25311 | FLJ41105 | KIAA0457 | SCZD9 | disrupted in schizophrenia 1 | DISC-1 interacts with RanBPM. | BIND | 12812986 |
| DYRK1B | MIRK | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B | - | HPRD,BioGRID | 14500717 |
| FMR1 | FMRP | FRAXA | MGC87458 | POF | POF1 | fragile X mental retardation 1 | FMR1 (FMRP) interacts with RANBP9 (RanBPM). | BIND | 15381419 |
| HIPK2 | DKFZp686K02111 | FLJ23711 | PRO0593 | homeodomain interacting protein kinase 2 | The nuclear protein kinase HIPK2 interacts with the Ran binding protein RanBPM | BIND | 12220523 |
| HIPK2 | DKFZp686K02111 | FLJ23711 | PRO0593 | homeodomain interacting protein kinase 2 | - | HPRD,BioGRID | 12220523 |
| ITGB2 | CD18 | LAD | LCAMB | LFA-1 | MAC-1 | MF17 | MFI7 | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) | RanBPM interacts with beta-2 integrin LFA-1. | BIND | 14722085 |
| L1CAM | CAML1 | CD171 | HSAS | HSAS1 | MASA | MIC5 | N-CAML1 | S10 | SPG1 | L1 cell adhesion molecule | L1CAM interacts with RanBPM. | BIND | 16000162 |
| MET | AUTS9 | HGFR | RCCP2 | c-Met | met proto-oncogene (hepatocyte growth factor receptor) | - | HPRD,BioGRID | 12147692 |
| MPHOSPH8 | FLJ35237 | HSMPP8 | TWA3 | mpp8 | M-phase phosphoprotein 8 | - | HPRD,BioGRID | 12559565 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | Affinity Capture-Western Two-hybrid | BioGRID | 12361945 |
| RAN | ARA24 | Gsp1 | TC4 | RAN, member RAS oncogene family | - | HPRD,BioGRID | 9817760 |
| RNPS1 | E5.1 | MGC117332 | RNA binding protein S1, serine-rich domain | Affinity Capture-MS | BioGRID | 17353931 |
| S100A7 | PSOR1 | S100A7c | S100 calcium binding protein A7 | - | HPRD,BioGRID | 12421467 |
| UCHL1 | PARK5 | PGP9.5 | Uch-L1 | ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) | - | HPRD,BioGRID | 12082530 |
| USP11 | UHX1 | ubiquitin specific peptidase 11 | - | HPRD,BioGRID | 12084015 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID MET PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID AR PATHWAY | 61 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME L1CAM INTERACTIONS | 86 | 62 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS UP | 112 | 68 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR DN | 214 | 133 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| DEN INTERACT WITH LCA5 | 26 | 21 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 6P24 P22 AMPLICON | 21 | 17 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL AND BRAIN QTL TRANS | 185 | 114 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| JIANG VHL TARGETS | 138 | 91 | All SZGR 2.0 genes in this pathway |
| JI RESPONSE TO FSH DN | 58 | 43 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR UP | 55 | 41 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| ABRAMSON INTERACT WITH AIRE | 45 | 33 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 507 | 514 | 1A,m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-144 | 508 | 514 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-200bc/429 | 681 | 688 | 1A,m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-208 | 753 | 759 | 1A | hsa-miR-208 | AUAAGACGAGCAAAAAGCUUGU |
| miR-214 | 64 | 70 | 1A | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-25/32/92/363/367 | 810 | 816 | 1A | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-26 | 517 | 523 | 1A | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-30-5p | 137 | 143 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-323 | 501 | 507 | m8 | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-410 | 718 | 724 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-411 | 748 | 754 | m8 | hsa-miR-411 | AACACGGUCCACUAACCCUCAGU |
| miR-496 | 375 | 381 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-499 | 753 | 759 | 1A | hsa-miR-499 | UUAAGACUUGCAGUGAUGUUUAA |
| miR-505 | 478 | 484 | m8 | hsa-miR-505 | GUCAACACUUGCUGGUUUCCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.