Gene Page: YAP1
Summary ?
| GeneID | 10413 |
| Symbol | YAP1 |
| Synonyms | COB1|YAP|YAP2|YAP65|YKI |
| Description | Yes associated protein 1 |
| Reference | MIM:606608|HGNC:HGNC:16262|Ensembl:ENSG00000137693|HPRD:09424|Vega:OTTHUMG00000167322 |
| Gene type | protein-coding |
| Map location | 11q13 |
| Pascal p-value | 0.276 |
| Sherlock p-value | 0.096 |
| Fetal beta | 1.1 |
| DMG | 2 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| Expression | Meta-analysis of gene expression | P value: 3.166 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.6564 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01442799 | 11 | 101981050 | YAP1 | -0.026 | 0.28 | DMG:Nishioka_2013 | |
| cg01442799 | 11 | 101981050 | YAP1 | 3.24E-8 | -0.008 | 9.74E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
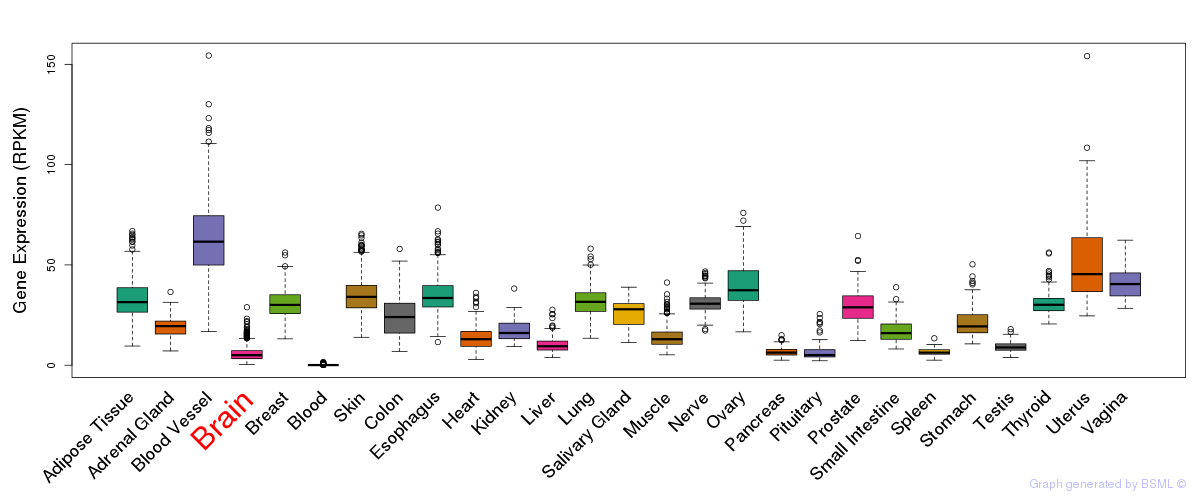
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CGGBP1 | 0.90 | 0.91 |
| KBTBD7 | 0.89 | 0.89 |
| DLSTP | 0.89 | 0.89 |
| LEPROTL1 | 0.88 | 0.90 |
| ASB7 | 0.88 | 0.88 |
| ADNP2 | 0.88 | 0.89 |
| PIP5K1A | 0.88 | 0.89 |
| DCTN4 | 0.88 | 0.88 |
| MINPP1 | 0.88 | 0.88 |
| PTPN9 | 0.88 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.71 | -0.80 |
| AF347015.31 | -0.70 | -0.79 |
| AF347015.27 | -0.68 | -0.77 |
| AF347015.8 | -0.68 | -0.80 |
| MT-CYB | -0.68 | -0.78 |
| AF347015.33 | -0.68 | -0.76 |
| FXYD1 | -0.68 | -0.78 |
| HSD17B14 | -0.67 | -0.73 |
| HIGD1B | -0.66 | -0.77 |
| AF347015.21 | -0.66 | -0.81 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 7644498 | |
| GO:0016563 | transcription activator activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005667 | transcription factor complex | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| DENND3 | KIAA0870 | DENN/MADD domain containing 3 | Two-hybrid | BioGRID | 11278422 |
| ERBB4 | HER4 | MGC138404 | p180erbB4 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) | - | HPRD,BioGRID | 15023535 |
| KIF5A | D12S1889 | MY050 | NKHC | SPG10 | kinesin family member 5A | Two-hybrid | BioGRID | 11278422 |
| NFE2 | NF-E2 | p45 | nuclear factor (erythroid-derived 2), 45kDa | p45 interacts with hYAP. | BIND | 9305852 |
| RUNX1 | AML1 | AML1-EVI-1 | AMLCR1 | CBFA2 | EVI-1 | PEBP2aB | runt-related transcription factor 1 | - | HPRD,BioGRID | 10228168 |
| RUNX2 | AML3 | CBFA1 | CCD | CCD1 | MGC120022 | MGC120023 | OSF2 | PEA2aA | PEBP2A1 | PEBP2A2 | PEBP2aA | PEBP2aA1 | runt-related transcription factor 2 | - | HPRD,BioGRID | 10228168 |
| RUNX3 | AML2 | CBFA3 | FLJ34510 | MGC16070 | PEBP2aC | runt-related transcription factor 3 | Reconstituted Complex | BioGRID | 10228168 |
| SLC9A3R1 | EBP50 | NHERF | NHERF1 | NPHLOP2 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 | - | HPRD,BioGRID | 10562288 |
| SMAD7 | CRCS3 | FLJ16482 | MADH7 | MADH8 | SMAD family member 7 | - | HPRD,BioGRID | 12118366 |
| TEAD1 | AA | REF1 | TCF13 | TEF-1 | TEA domain family member 1 (SV40 transcriptional enhancer factor) | - | HPRD | 11358867 |
| TEAD2 | ETF | TEF-4 | TEF4 | TEA domain family member 2 | - | HPRD | 11358867 |
| TEAD3 | DTEF-1 | ETFR-1 | TEAD5 | TEF-5 | TEF5 | TEA domain family member 3 | - | HPRD | 11358867 |
| TEAD4 | EFTR-2 | MGC9014 | RTEF1 | TCF13L1 | TEF-3 | TEFR-1 | hRTEF-1B | TEA domain family member 4 | - | HPRD | 11358867 |
| TP53BP2 | 53BP2 | ASPP2 | BBP | PPP1R13A | p53BP2 | tumor protein p53 binding protein, 2 | - | HPRD,BioGRID | 11278422 |
| TP63 | AIS | B(p51A) | B(p51B) | EEC3 | KET | LMS | NBP | OFC8 | RHS | SHFM4 | TP53CP | TP53L | TP73L | p40 | p51 | p53CP | p63 | p73H | p73L | tumor protein p63 | Reconstituted Complex | BioGRID | 11278685 |
| TP73 | P73 | tumor protein p73 | - | HPRD,BioGRID | 11278685 |
| TP73 | P73 | tumor protein p73 | p73-alpha interacts with YAP. | BIND | 15893728 |
| WBP1 | MGC15305 | WBP-1 | WW domain binding protein 1 | in vitro in vivo Protein-peptide | BioGRID | 7644498 |11604498 |
| WBP2 | MGC18269 | WBP-2 | WW domain binding protein 2 | in vitro | BioGRID | 7644498 |
| YES1 | HsT441 | P61-YES | Yes | c-yes | v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 | - | HPRD | 10562288 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | - | HPRD,BioGRID | 12535517 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID ERBB4 PATHWAY | 38 | 32 | All SZGR 2.0 genes in this pathway |
| PID P73PATHWAY | 79 | 59 | All SZGR 2.0 genes in this pathway |
| PID DELTA NP63 PATHWAY | 47 | 34 | All SZGR 2.0 genes in this pathway |
| PID TGFBR PATHWAY | 55 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR SIGNALING BY ERBB4 | 38 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME PPARA ACTIVATES GENE EXPRESSION | 104 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY HIPPO | 22 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | 24 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | 168 | 115 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA UP | 205 | 140 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| SUZUKI AMPLIFIED IN ORAL CANCER | 16 | 12 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| SNIJDERS AMPLIFIED IN HEAD AND NECK TUMORS | 37 | 27 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE UP | 283 | 177 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS UP | 206 | 118 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR UP | 174 | 96 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 DN | 228 | 114 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA MESENCHYMAL | 216 | 130 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-129-5p | 1993 | 1999 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-21 | 1714 | 1721 | 1A,m8 | hsa-miR-21brain | UAGCUUAUCAGACUGAUGUUGA |
| hsa-miR-590 | GAGCUUAUUCAUAAAAGUGCAG | ||||
| miR-299-3p | 652 | 658 | 1A | hsa-miR-299-3p | UAUGUGGGAUGGUAAACCGCUU |
| miR-299-5p | 2776 | 2782 | 1A | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
| miR-375 | 1683 | 1689 | 1A | hsa-miR-375 | UUUGUUCGUUCGGCUCGCGUGA |
| miR-9 | 1663 | 1669 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.