Gene Page: HTATIP2
Summary ?
| GeneID | 10553 |
| Symbol | HTATIP2 |
| Synonyms | CC3|SDR44U1|TIP30 |
| Description | HIV-1 Tat interactive protein 2 |
| Reference | MIM:605628|HGNC:HGNC:16637|Ensembl:ENSG00000109854|HPRD:09288|Vega:OTTHUMG00000166015 |
| Gene type | protein-coding |
| Map location | 11p15.1 |
| Pascal p-value | 0.287 |
| Sherlock p-value | 0.109 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Cortex Hippocampus Nucleus accumbens basal ganglia Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg25533556 | 11 | 20385280 | HTATIP2 | 3.95E-6 | -0.74 | 0.009 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11025524 | 11 | 20379276 | HTATIP2 | ENSG00000109854.9 | 9.297E-12 | 0 | -5955 | gtex_brain_putamen_basal |
| rs11025525 | 11 | 20379963 | HTATIP2 | ENSG00000109854.9 | 1.187E-11 | 0 | -5268 | gtex_brain_putamen_basal |
| rs4757940 | 11 | 20382453 | HTATIP2 | ENSG00000109854.9 | 1.367E-11 | 0 | -2778 | gtex_brain_putamen_basal |
| rs12360593 | 11 | 20384003 | HTATIP2 | ENSG00000109854.9 | 1.203E-11 | 0 | -1228 | gtex_brain_putamen_basal |
| rs397753461 | 11 | 20384886 | HTATIP2 | ENSG00000109854.9 | 3.251E-11 | 0 | -345 | gtex_brain_putamen_basal |
| rs10437608 | 11 | 20385606 | HTATIP2 | ENSG00000109854.9 | 1.637E-11 | 0 | 375 | gtex_brain_putamen_basal |
| rs10833312 | 11 | 20386301 | HTATIP2 | ENSG00000109854.9 | 5.031E-11 | 0 | 1070 | gtex_brain_putamen_basal |
| rs10437583 | 11 | 20386761 | HTATIP2 | ENSG00000109854.9 | 4.793E-11 | 0 | 1530 | gtex_brain_putamen_basal |
| rs11025535 | 11 | 20387062 | HTATIP2 | ENSG00000109854.9 | 3.709E-9 | 0 | 1831 | gtex_brain_putamen_basal |
| rs10833313 | 11 | 20387375 | HTATIP2 | ENSG00000109854.9 | 3.954E-11 | 0 | 2144 | gtex_brain_putamen_basal |
| rs7925136 | 11 | 20387798 | HTATIP2 | ENSG00000109854.9 | 4.687E-9 | 0 | 2567 | gtex_brain_putamen_basal |
| rs7928503 | 11 | 20388097 | HTATIP2 | ENSG00000109854.9 | 2.638E-9 | 0 | 2866 | gtex_brain_putamen_basal |
| rs7103934 | 11 | 20389743 | HTATIP2 | ENSG00000109854.9 | 1.026E-9 | 0 | 4512 | gtex_brain_putamen_basal |
| rs7112213 | 11 | 20391426 | HTATIP2 | ENSG00000109854.9 | 9.743E-9 | 0 | 6195 | gtex_brain_putamen_basal |
| rs11607982 | 11 | 20395162 | HTATIP2 | ENSG00000109854.9 | 1.192E-6 | 0 | 9931 | gtex_brain_putamen_basal |
| rs7937333 | 11 | 20499112 | HTATIP2 | ENSG00000109854.9 | 7.613E-7 | 0 | 113881 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
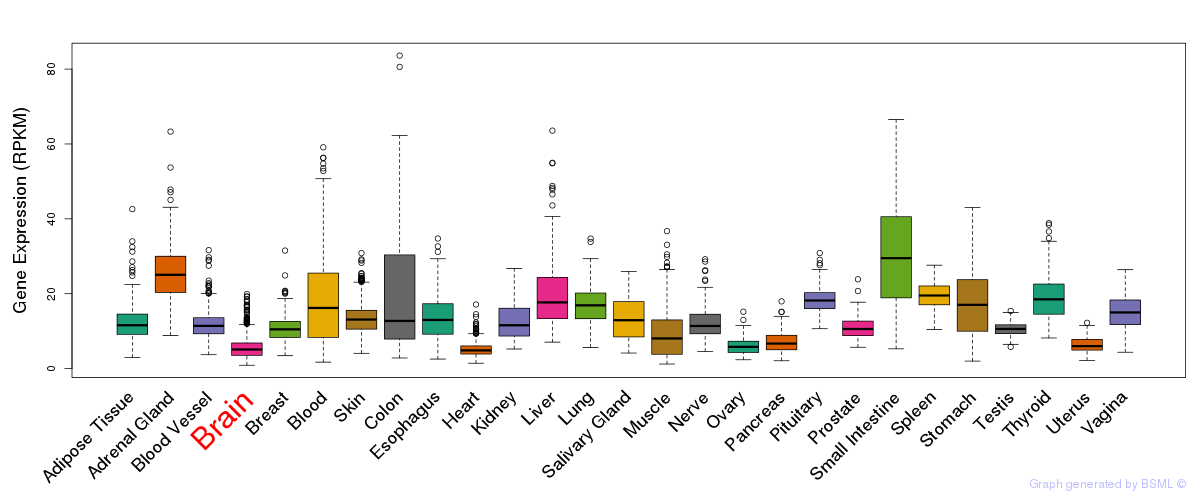
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| KIM RESPONSE TO TSA AND DECITABINE UP | 129 | 73 | All SZGR 2.0 genes in this pathway |
| SAMOLS TARGETS OF KHSV MIRNAS DN | 62 | 35 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS UP | 306 | 188 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F DN | 33 | 24 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA LOH REGIONS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| MANTOVANI VIRAL GPCR SIGNALING UP | 86 | 54 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE UP | 77 | 60 | All SZGR 2.0 genes in this pathway |
| FRIDMAN IMMORTALIZATION DN | 34 | 24 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES CORE NINE CORRELATED | 100 | 68 | All SZGR 2.0 genes in this pathway |
| MORI PLASMA CELL UP | 51 | 29 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC UP | 54 | 30 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA UP | 61 | 40 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER ACOX1 UP | 64 | 40 | All SZGR 2.0 genes in this pathway |
| UEDA PERIFERAL CLOCK | 169 | 111 | All SZGR 2.0 genes in this pathway |
| SMITH LIVER CANCER | 45 | 27 | All SZGR 2.0 genes in this pathway |
| GERY CEBP TARGETS | 126 | 90 | All SZGR 2.0 genes in this pathway |
| GALE APL WITH FLT3 MUTATED UP | 56 | 35 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS MATURE CELL | 293 | 160 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP | 181 | 106 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 COMPLETE | 227 | 151 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS UP | 266 | 142 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |