Gene Page: CELF2
Summary ?
| GeneID | 10659 |
| Symbol | CELF2 |
| Synonyms | BRUNOL3|CUGBP2|ETR-3|ETR3|NAPOR |
| Description | CUGBP, Elav-like family member 2 |
| Reference | MIM:602538|HGNC:HGNC:2550|Ensembl:ENSG00000048740|HPRD:03964|Vega:OTTHUMG00000017668 |
| Gene type | protein-coding |
| Map location | 10p13 |
| Pascal p-value | 0.002 |
| Sherlock p-value | 0.617 |
| TADA p-value | 0.001 |
| Fetal beta | 1.193 |
| eGene | Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Xu_2012 | Whole Exome Sequencing analysis | De novo mutations of 4 genes were identified by exome sequencing of 795 samples in this study |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CELF2 | chr10 | 11356223 | T | N | NM_001025076 NM_001025076 NM_001025077 NM_001025077 NM_001083591 NM_006561 NM_006561 | . . . . p.335G>? . . | intronic splice-donor-in2 intronic splice-donor-in2 exonic-unknown intronic splice-donor-in2 | Schizophrenia | DNM:Xu_2012 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10508433 | chr10 | 11869520 | CELF2 | 10659 | 0.06 | cis | ||
| rs10868543 | chr9 | 89852686 | CELF2 | 10659 | 0.15 | trans |
Section II. Transcriptome annotation
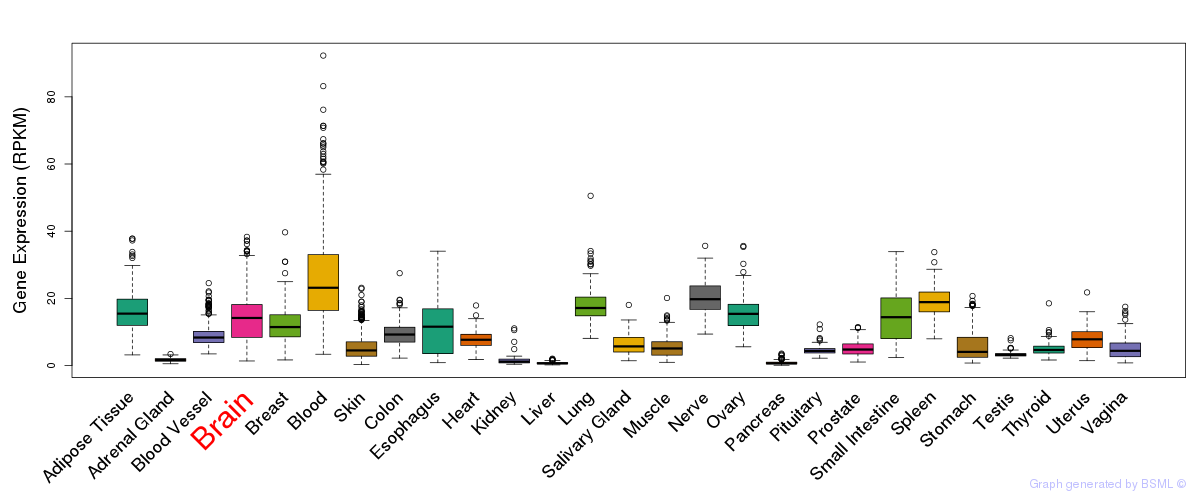
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GFOD2 | 0.87 | 0.88 |
| DGCR14 | 0.87 | 0.90 |
| CARM1 | 0.87 | 0.90 |
| CTBP1 | 0.87 | 0.89 |
| RAB11B | 0.87 | 0.89 |
| RAB35 | 0.86 | 0.89 |
| BAT3 | 0.86 | 0.89 |
| PRCC | 0.86 | 0.89 |
| G6PD | 0.86 | 0.89 |
| C7orf26 | 0.86 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.77 | -0.84 |
| AF347015.31 | -0.77 | -0.83 |
| MT-CYB | -0.74 | -0.81 |
| AF347015.33 | -0.73 | -0.80 |
| AF347015.8 | -0.73 | -0.82 |
| AF347015.27 | -0.72 | -0.81 |
| AF347015.21 | -0.70 | -0.83 |
| NOSTRIN | -0.70 | -0.79 |
| AF347015.2 | -0.69 | -0.79 |
| AF347015.15 | -0.69 | -0.79 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003723 | RNA binding | TAS | 9887331 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006397 | mRNA processing | IEA | - | |
| GO:0008016 | regulation of heart contraction | TAS | 9887331 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| WINTER HYPOXIA DN | 52 | 30 | All SZGR 2.0 genes in this pathway |
| LU TUMOR VASCULATURE DN | 13 | 6 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST NORMAL DUCTAL VS LOBULAR UP | 68 | 40 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER GRADES 1 2 DN | 67 | 43 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2A DN | 141 | 84 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 AND TP63 TARGETS | 205 | 145 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS DN | 124 | 79 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING UP | 108 | 69 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING VIA SMAD4 UP | 108 | 66 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE DN | 165 | 104 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| KENNY CTNNB1 TARGETS UP | 50 | 30 | All SZGR 2.0 genes in this pathway |
| PARK HSC VS MULTIPOTENT PROGENITORS UP | 19 | 14 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK UP | 87 | 58 | All SZGR 2.0 genes in this pathway |
| ASTON MAJOR DEPRESSIVE DISORDER UP | 49 | 36 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP | 207 | 145 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA UP | 98 | 64 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAINE REWARD 5D | 79 | 62 | All SZGR 2.0 genes in this pathway |
| LINDVALL IMMORTALIZED BY TERT DN | 80 | 56 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| KRASNOSELSKAYA ILF3 TARGETS DN | 46 | 38 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| ZHENG FOXP3 TARGETS IN THYMUS UP | 196 | 137 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION DN | 281 | 179 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| LEE DIFFERENTIATING T LYMPHOCYTE | 200 | 115 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G5 DN | 27 | 17 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 TARGETS UP | 66 | 43 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS DN | 207 | 139 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| GREGORY SYNTHETIC LETHAL WITH IMATINIB | 145 | 83 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 UP | 134 | 85 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS UP | 116 | 87 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 10HR DN | 30 | 25 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-10 | 717 | 723 | m8 | hsa-miR-10a | UACCCUGUAGAUCCGAAUUUGUG |
| hsa-miR-10b | UACCCUGUAGAACCGAAUUUGU | ||||
| miR-101 | 2121 | 2127 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-103/107 | 1612 | 1618 | 1A | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-129-5p | 2250 | 2256 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC | ||||
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC | ||||
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-132/212 | 7206 | 7212 | m8 | hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC |
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| miR-134 | 5102 | 5108 | m8 | hsa-miR-134brain | UGUGACUGGUUGACCAGAGGG |
| miR-137 | 1769 | 1775 | 1A | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-142-3p | 550 | 556 | 1A | hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA |
| miR-144 | 2121 | 2127 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG | ||||
| miR-145 | 1147 | 1153 | m8 | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-146 | 942 | 948 | m8 | hsa-miR-146a | UGAGAACUGAAUUCCAUGGGUU |
| hsa-miR-146bbrain | UGAGAACUGAAUUCCAUAGGCU | ||||
| miR-153 | 7370 | 7376 | m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-155 | 5006 | 5012 | m8 | hsa-miR-155 | UUAAUGCUAAUCGUGAUAGGGG |
| miR-17-5p/20/93.mr/106/519.d | 5106 | 5112 | 1A | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-182 | 2442 | 2448 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-183 | 3480 | 3487 | 1A,m8 | hsa-miR-183 | UAUGGCACUGGUAGAAUUCACUG |
| miR-196 | 2014 | 2020 | 1A | hsa-miR-196a | UAGGUAGUUUCAUGUUGUUGG |
| hsa-miR-196b | UAGGUAGUUUCCUGUUGUUGG | ||||
| miR-199 | 628 | 634 | m8 | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-200bc/429 | 1724 | 1730 | 1A | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-208 | 5020 | 5026 | 1A | hsa-miR-208 | AUAAGACGAGCAAAAAGCUUGU |
| hsa-miR-208 | AUAAGACGAGCAAAAAGCUUGU | ||||
| miR-214 | 3461 | 3467 | m8 | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-216 | 4832 | 4838 | 1A | hsa-miR-216 | UAAUCUCAGCUGGCAACUGUG |
| miR-219 | 7177 | 7183 | 1A | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU | ||||
| miR-23 | 1598 | 1605 | 1A,m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC | ||||
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC | ||||
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-24 | 4746 | 4752 | 1A | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-26 | 1717 | 1723 | 1A | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC | ||||
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-299-5p | 5288 | 5294 | 1A | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
| miR-30-3p | 1629 | 1635 | m8 | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-30-5p | 3776 | 3782 | m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-323 | 1598 | 1604 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| hsa-miR-323brain | GCACAUUACACGGUCGACCUCU | ||||
| hsa-miR-323brain | GCACAUUACACGGUCGACCUCU | ||||
| miR-324-3p | 1891 | 1897 | m8 | hsa-miR-324-3p | CCACUGCCCCAGGUGCUGCUGG |
| miR-326 | 2802 | 2808 | m8 | hsa-miR-326 | CCUCUGGGCCCUUCCUCCAG |
| miR-33 | 1262 | 1268 | 1A | hsa-miR-33 | GUGCAUUGUAGUUGCAUUG |
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| hsa-miR-33 | GUGCAUUGUAGUUGCAUUG | ||||
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| miR-339 | 4300 | 4306 | 1A | hsa-miR-339 | UCCCUGUCCUCCAGGAGCUCA |
| hsa-miR-339 | UCCCUGUCCUCCAGGAGCUCA | ||||
| miR-34/449 | 417 | 423 | m8 | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| miR-340 | 116 | 122 | m8 | hsa-miR-340 | UCCGUCUCAGUUACUUUAUAGCC |
| miR-342 | 2489 | 2495 | 1A | hsa-miR-342brain | UCUCACACAGAAAUCGCACCCGUC |
| miR-34b | 878 | 884 | m8 | hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
| hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG | ||||
| miR-361 | 7148 | 7154 | 1A | hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC |
| miR-376 | 1579 | 1585 | 1A | hsa-miR-376a | AUCAUAGAGGAAAAUCCACGU |
| hsa-miR-376b | AUCAUAGAGGAAAAUCCAUGUU | ||||
| miR-377 | 5253 | 5259 | m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-378 | 7197 | 7203 | 1A | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
| hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU | ||||
| miR-381 | 2531 | 2537 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU | ||||
| miR-384 | 992 | 998 | m8 | hsa-miR-384 | AUUCCUAGAAAUUGUUCAUA |
| miR-410 | 7409 | 7415 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-411 | 5207 | 5214 | 1A,m8 | hsa-miR-411 | AACACGGUCCACUAACCCUCAGU |
| miR-431 | 1624 | 1630 | m8 | hsa-miR-431 | UGUCUUGCAGGCCGUCAUGCA |
| miR-433-5p | 1010 | 1016 | m8 | hsa-miR-433-5p | UACGGUGAGCCUGUCAUUAUUC |
| miR-450 | 3689 | 3695 | 1A | hsa-miR-450 | UUUUUGCGAUGUGUUCCUAAUA |
| miR-451 | 119 | 125 | 1A | hsa-miR-451brain | AAACCGUUACCAUUACUGAGUUU |
| miR-452 | 359 | 365 | 1A | hsa-miR-452 | UGUUUGCAGAGGAAACUGAGAC |
| miR-486 | 2106 | 2113 | 1A,m8 | hsa-miR-486 | UCCUGUACUGAGCUGCCCCGAG |
| miR-493-5p | 2104 | 2110 | m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-494 | 754 | 760 | m8 | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
| miR-495 | 643 | 649 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU | ||||
| miR-499 | 5020 | 5026 | 1A | hsa-miR-499 | UUAAGACUUGCAGUGAUGUUUAA |
| hsa-miR-499 | UUAAGACUUGCAGUGAUGUUUAA | ||||
| miR-539 | 1934 | 1940 | 1A | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
| hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU | ||||
| miR-543 | 1792 | 1798 | m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 523 | 530 | 1A,m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
| miR-96 | 2441 | 2448 | 1A,m8 | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.