Gene Page: MAPRE2
Summary ?
| GeneID | 10982 |
| Symbol | MAPRE2 |
| Synonyms | CSCSC2|EB1|EB2|RP1 |
| Description | microtubule associated protein RP/EB family member 2 |
| Reference | MIM:605789|HGNC:HGNC:6891|Ensembl:ENSG00000166974|HPRD:16157|Vega:OTTHUMG00000132551 |
| Gene type | protein-coding |
| Map location | 18q12.1 |
| Pascal p-value | 0.331 |
| Sherlock p-value | 0.227 |
| Fetal beta | -0.298 |
| DMG | 2 (# studies) |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07020962 | 18 | 32621292 | MAPRE2 | -0.026 | 0.27 | DMG:Nishioka_2013 | |
| cg24692068 | 18 | 32557882 | MAPRE2 | 3.76E-8 | -0.009 | 1.09E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
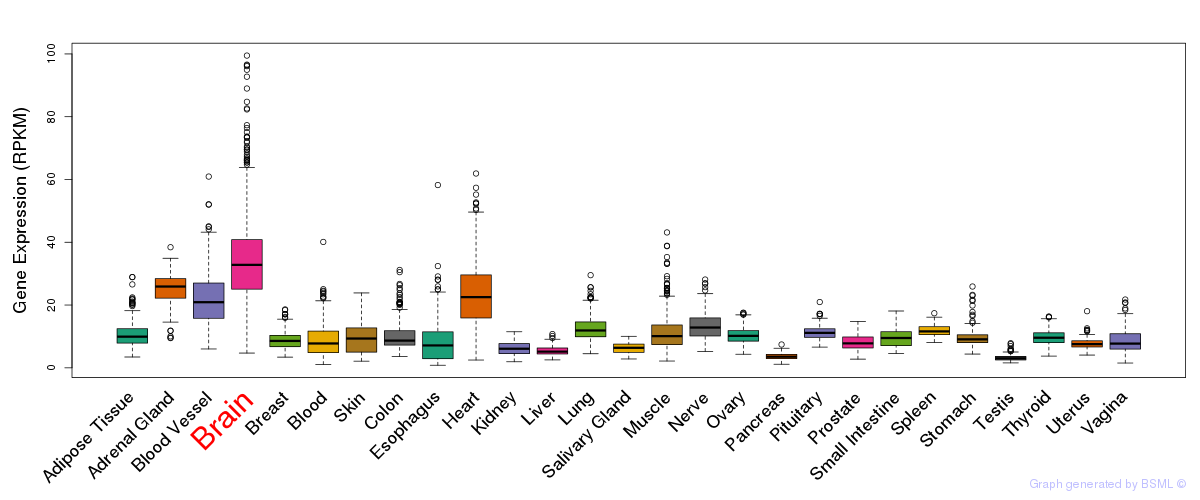
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TTLL7 | 0.86 | 0.79 |
| FAM73A | 0.86 | 0.79 |
| STRN | 0.84 | 0.82 |
| ARFGEF2 | 0.84 | 0.80 |
| DPP8 | 0.84 | 0.78 |
| NCOA7 | 0.84 | 0.81 |
| LANCL1 | 0.84 | 0.82 |
| PEG3AS | 0.83 | 0.85 |
| ZFP106 | 0.83 | 0.80 |
| RAPGEF5 | 0.83 | 0.72 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC006276.2 | -0.46 | -0.49 |
| EFEMP2 | -0.44 | -0.49 |
| TBC1D10A | -0.43 | -0.40 |
| C1orf61 | -0.42 | -0.54 |
| BCL7C | -0.42 | -0.50 |
| RAMP2 | -0.42 | -0.49 |
| NUDT8 | -0.42 | -0.49 |
| IRF7 | -0.40 | -0.51 |
| RAB34 | -0.39 | -0.47 |
| WDR86 | -0.39 | -0.39 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A UP | 191 | 128 | All SZGR 2.0 genes in this pathway |
| BARRIER CANCER RELAPSE NORMAL SAMPLE UP | 32 | 21 | All SZGR 2.0 genes in this pathway |
| SEITZ NEOPLASTIC TRANSFORMATION BY 8P DELETION UP | 73 | 47 | All SZGR 2.0 genes in this pathway |
| WOOD EBV EBNA1 TARGETS UP | 110 | 71 | All SZGR 2.0 genes in this pathway |
| SCIAN CELL CYCLE TARGETS OF TP53 AND TP73 DN | 22 | 14 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| LEE TARGETS OF PTCH1 AND SUFU DN | 83 | 69 | All SZGR 2.0 genes in this pathway |
| KLEIN PRIMARY EFFUSION LYMPHOMA DN | 58 | 42 | All SZGR 2.0 genes in this pathway |
| PEART HDAC PROLIFERATION CLUSTER UP | 57 | 35 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS B LYMPHOCYTE DN | 38 | 25 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER METASTASIS DN | 121 | 65 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| CHEN LVAD SUPPORT OF FAILING HEART UP | 103 | 69 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS INTERMEDIATE PROGENITOR | 149 | 84 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 CORE DIRECT TARGETS | 19 | 14 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 2G | 171 | 96 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE UP | 126 | 92 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN | 240 | 153 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 TARGETS UP | 66 | 43 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS DN | 87 | 66 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN DN | 88 | 68 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 DN | 157 | 106 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |