Gene Page: WWP2
Summary ?
| GeneID | 11060 |
| Symbol | WWP2 |
| Synonyms | AIP2|WWp2-like |
| Description | WW domain containing E3 ubiquitin protein ligase 2 |
| Reference | MIM:602308|HGNC:HGNC:16804|Ensembl:ENSG00000198373|HPRD:03812|Vega:OTTHUMG00000137573 |
| Gene type | protein-coding |
| Map location | 16q22.1 |
| Pascal p-value | 0.241 |
| Sherlock p-value | 0.599 |
| Fetal beta | -0.297 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg16516248 | 16 | 69958626 | WWP2 | 3.795E-4 | 0.425 | 0.043 | DMG:Wockner_2014 |
| cg02712546 | 16 | 69969166 | WWP2 | 4.41E-4 | 0.424 | 0.045 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
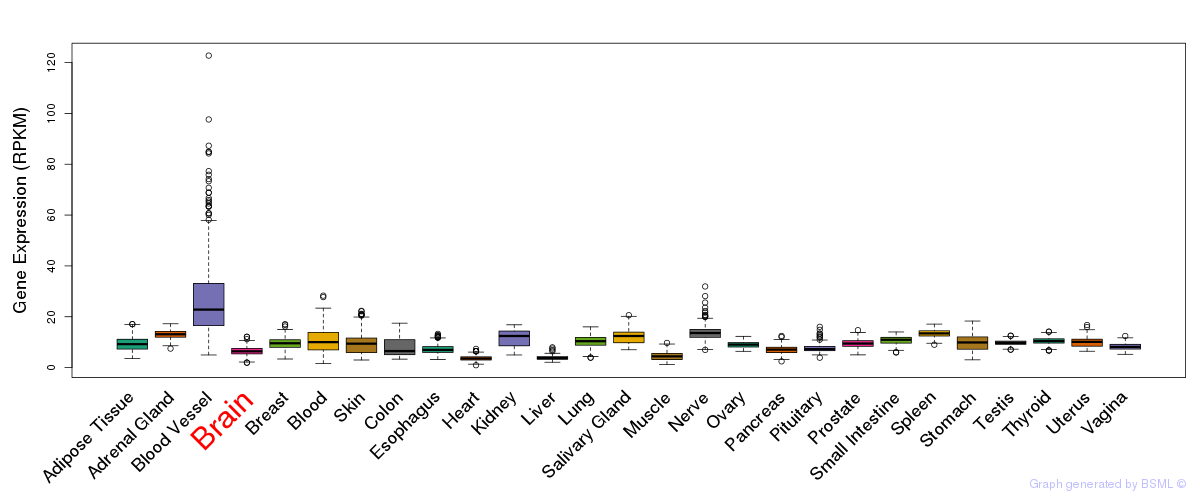
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATN1 | B37 | D12S755E | DRPLA | NOD | atrophin 1 | Atrophin-1 interacts with AIP2. | BIND | 9647693 |
| ATN1 | B37 | D12S755E | DRPLA | NOD | atrophin 1 | - | HPRD,BioGRID | 9647693 |
| CLCN5 | CLC5 | CLCK2 | DENTS | NPHL1 | NPHL2 | XLRH | XRN | hCIC-K2 | hClC-K2 | chloride channel 5 | - | HPRD | 9169421 |
| CPSF6 | CFIM | CFIM68 | HPBRII-4 | HPBRII-7 | cleavage and polyadenylation specific factor 6, 68kDa | Two-hybrid | BioGRID | 16189514 |
| DAG1 | 156DAG | A3a | AGRNR | DAG | dystroglycan 1 (dystrophin-associated glycoprotein 1) | - | HPRD | 9169421 |
| DIDO1 | BYE1 | C20orf158 | DATF1 | DIDO2 | DIDO3 | DIO-1 | DIO1 | DKFZp434P1115 | FLJ11265 | KIAA0333 | MGC16140 | dJ885L7.8 | death inducer-obliterator 1 | Two-hybrid | BioGRID | 16189514 |
| EWSR1 | EWS | Ewing sarcoma breakpoint region 1 | Two-hybrid | BioGRID | 16189514 |
| IL2RG | CD132 | IMD4 | SCIDX | SCIDX1 | interleukin 2 receptor, gamma (severe combined immunodeficiency) | - | HPRD | 9169421 |
| IL6R | CD126 | IL-6R-1 | IL-6R-alpha | IL6RA | MGC104991 | interleukin 6 receptor | - | HPRD | 9169421 |
| NDFIP2 | FLJ25842 | KIAA1165 | N4wbp5a | Nedd4 family interacting protein 2 | - | HPRD | 15252135 |
| PDLIM7 | LMP1 | PDZ and LIM domain 7 (enigma) | Two-hybrid | BioGRID | 16189514 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | - | HPRD | 9169421 |
| SCNN1A | ENaCa | ENaCalpha | FLJ21883 | SCNEA | SCNN1 | sodium channel, nonvoltage-gated 1 alpha | - | HPRD,BioGRID | 12167593 |
| SCNN1B | ENaCb | ENaCbeta | SCNEB | sodium channel, nonvoltage-gated 1, beta | - | HPRD,BioGRID | 12167593 |
| SCNN1G | ENaCg | ENaCgamma | PHA1 | SCNEG | sodium channel, nonvoltage-gated 1, gamma | - | HPRD,BioGRID | 12167593 |
| SF1 | D11S636 | ZFM1 | ZNF162 | splicing factor 1 | Two-hybrid | BioGRID | 16189514 |
| TFAP2A | AP-2 | AP-2alpha | AP2TF | BOFS | TFAP2 | transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) | - | HPRD | 9169421 |
| TP53BP2 | 53BP2 | ASPP2 | BBP | PPP1R13A | p53BP2 | tumor protein p53 binding protein, 2 | - | HPRD | 9169421 |
| VASP | - | vasodilator-stimulated phosphoprotein | Two-hybrid | BioGRID | 16189514 |
| WBP1 | MGC15305 | WBP-1 | WW domain binding protein 1 | - | HPRD,BioGRID | 9169421 |
| WBP2 | MGC18269 | WBP-2 | WW domain binding protein 2 | Protein-peptide | BioGRID | 9169421 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG UBIQUITIN MEDIATED PROTEOLYSIS | 138 | 98 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS DN | 142 | 95 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 DN | 153 | 100 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION DN | 100 | 64 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 1 | 45 | 27 | All SZGR 2.0 genes in this pathway |
| SILIGAN BOUND BY EWS FLT1 FUSION | 47 | 34 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| WOTTON RUNX TARGETS UP | 21 | 14 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE UP | 157 | 105 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN UP | 181 | 112 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| NIELSEN GIST | 98 | 66 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| VANOEVELEN MYOGENESIS SIN3A TARGETS | 220 | 133 | All SZGR 2.0 genes in this pathway |