Gene Page: DNAJB4
Summary ?
| GeneID | 11080 |
| Symbol | DNAJB4 |
| Synonyms | DNAJW|DjB4|HLJ1 |
| Description | DnaJ heat shock protein family (Hsp40) member B4 |
| Reference | MIM:611327|HGNC:HGNC:14886|HPRD:07486| |
| Gene type | protein-coding |
| Map location | 1p31.1 |
| Pascal p-value | 0.047 |
| Sherlock p-value | 0.543 |
| Fetal beta | -0.834 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.02692 | |
| Expression | Meta-analysis of gene expression | P value: 1.445 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg25846022 | 1 | 78444909 | DNAJB4 | 6.39E-10 | -0.011 | 9.42E-7 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1879328 | chr3 | 46568156 | DNAJB4 | 11080 | 0.15 | trans | ||
| rs13350793 | 0 | DNAJB4 | 11080 | 0.18 | trans | |||
| rs1009417 | chr5 | 74568133 | DNAJB4 | 11080 | 0.1 | trans | ||
| rs12660028 | chr5 | 74605501 | DNAJB4 | 11080 | 0.04 | trans | ||
| rs1423526 | chr5 | 74606172 | DNAJB4 | 11080 | 0.04 | trans | ||
| rs6877840 | chr5 | 74611523 | DNAJB4 | 11080 | 0.15 | trans | ||
| rs10848708 | chr12 | 2931451 | DNAJB4 | 11080 | 0.06 | trans | ||
| rs2286601 | chr12 | 2932176 | DNAJB4 | 11080 | 0.14 | trans | ||
| rs8120542 | chr20 | 24395092 | DNAJB4 | 11080 | 0.05 | trans | ||
| rs8114637 | chr20 | 24395207 | DNAJB4 | 11080 | 0.05 | trans | ||
| rs4536719 | chr20 | 24397114 | DNAJB4 | 11080 | 0.17 | trans | ||
| rs9974024 | chr20 | 24397274 | DNAJB4 | 11080 | 0.05 | trans | ||
| rs6515493 | chr20 | 24401635 | DNAJB4 | 11080 | 0.05 | trans | ||
| rs6049703 | chr20 | 24432425 | DNAJB4 | 11080 | 0.14 | trans |
Section II. Transcriptome annotation
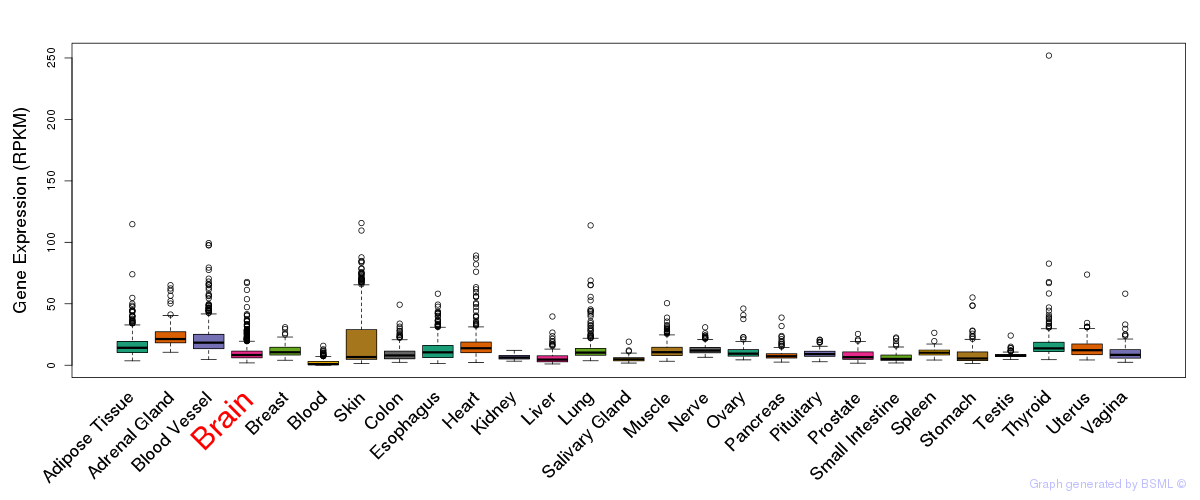
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GPRC5B | 0.94 | 0.88 |
| LDB3 | 0.93 | 0.91 |
| C10orf90 | 0.93 | 0.94 |
| EFHD1 | 0.93 | 0.93 |
| C5orf4 | 0.92 | 0.91 |
| ERBB3 | 0.92 | 0.93 |
| C11orf9 | 0.91 | 0.91 |
| AC022100.1 | 0.91 | 0.87 |
| PLEKHH1 | 0.91 | 0.78 |
| ATPGD1 | 0.91 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MED19 | -0.60 | -0.76 |
| NKIRAS2 | -0.60 | -0.68 |
| NR2C2AP | -0.60 | -0.78 |
| POLB | -0.59 | -0.78 |
| TRNAU1AP | -0.59 | -0.74 |
| HN1 | -0.58 | -0.73 |
| DUSP12 | -0.58 | -0.73 |
| ALKBH2 | -0.58 | -0.78 |
| ING4 | -0.58 | -0.73 |
| IFT52 | -0.58 | -0.75 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0031072 | heat shock protein binding | IEA | - | |
| GO:0051082 | unfolded protein binding | TAS | 9546042 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006457 | protein folding | IEA | - | |
| GO:0006986 | response to unfolded protein | TAS | 9546042 | |
| GO:0009408 | response to heat | TAS | 9546042 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP | 164 | 122 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 5 | 147 | 89 | All SZGR 2.0 genes in this pathway |
| DAUER STAT3 TARGETS DN | 50 | 34 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE | 217 | 143 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A UP | 142 | 104 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| SANA TNF SIGNALING DN | 90 | 57 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP | 205 | 126 | All SZGR 2.0 genes in this pathway |
| ADDYA ERYTHROID DIFFERENTIATION BY HEMIN | 73 | 47 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 2HR | 51 | 36 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 1 | 33 | 24 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 TARGETS STIMULATED DN | 11 | 9 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION DN | 105 | 67 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION DN | 281 | 179 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| PODAR RESPONSE TO ADAPHOSTIN UP | 147 | 98 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D UP | 139 | 95 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE S | 162 | 86 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 PARTIAL | 160 | 106 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR UP | 166 | 97 | All SZGR 2.0 genes in this pathway |
| GHANDHI BYSTANDER IRRADIATION UP | 86 | 54 | All SZGR 2.0 genes in this pathway |
| WARTERS RESPONSE TO IR SKIN | 83 | 44 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-24* | 151 | 157 | 1A | hsa-miR-189 | GUGCCUACUGAGCUGAUAUCAGU |
| miR-30-3p | 36 | 43 | 1A,m8 | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-539 | 213 | 219 | m8 | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.