Gene Page: CHRNB2
Summary ?
| GeneID | 1141 |
| Symbol | CHRNB2 |
| Synonyms | EFNL3|nAChRB2 |
| Description | cholinergic receptor nicotinic beta 2 subunit |
| Reference | MIM:118507|HGNC:HGNC:1962|Ensembl:ENSG00000160716|HPRD:00335|Vega:OTTHUMG00000037262 |
| Gene type | protein-coding |
| Map location | 1q21.3 |
| Fetal beta | 0.009 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere |
| Support | LIGAND GATED ION SIGNALING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 11 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17887936 | 1 | 154540354 | CHRNB2 | 2.7E-8 | -0.014 | 8.55E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
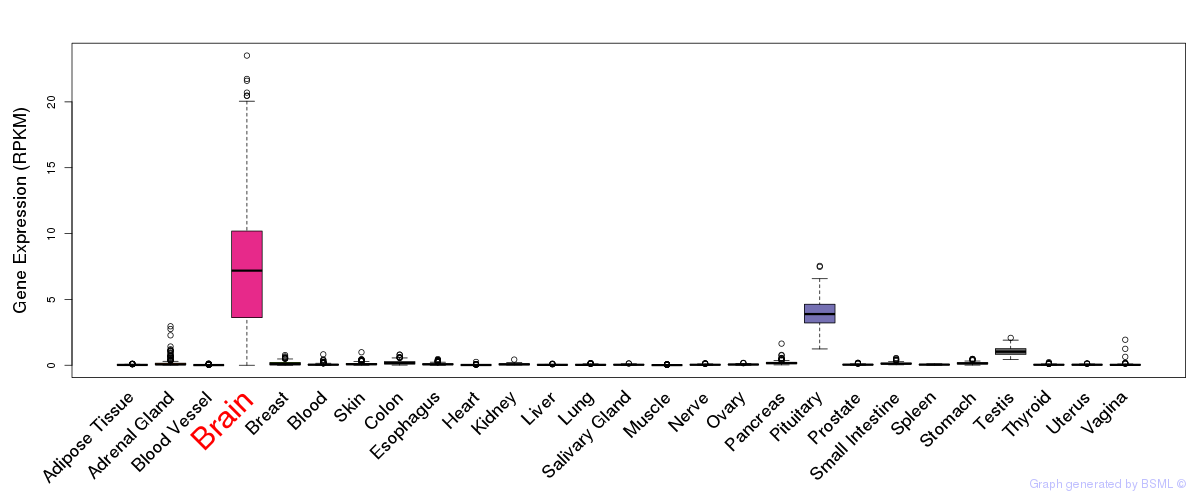
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SSBP3 | 0.73 | 0.75 |
| SORCS1 | 0.70 | 0.71 |
| KLHL32 | 0.70 | 0.67 |
| NXPH3 | 0.69 | 0.62 |
| EMX1 | 0.68 | 0.75 |
| AC127496.3 | 0.65 | 0.64 |
| TIAM2 | 0.65 | 0.69 |
| HS3ST4 | 0.65 | 0.67 |
| RGS6 | 0.63 | 0.66 |
| RP1-21O18.1 | 0.63 | 0.66 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CCDC80 | -0.48 | -0.61 |
| RHOC | -0.48 | -0.57 |
| S100A16 | -0.46 | -0.55 |
| TMEM51 | -0.46 | -0.54 |
| BDH2 | -0.45 | -0.52 |
| APOE | -0.45 | -0.55 |
| NMI | -0.45 | -0.53 |
| DBI | -0.45 | -0.56 |
| HSD17B14 | -0.45 | -0.51 |
| SERPINB6 | -0.45 | -0.48 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0015464 | acetylcholine receptor activity | IDA | Neurotransmitter (GO term level: 6) | 8906617 |
| GO:0004889 | nicotinic acetylcholine-activated cation-selective channel activity | IDA | 8906617 | |
| GO:0005515 | protein binding | ISS | - | |
| GO:0005216 | ion channel activity | IEA | - | |
| GO:0005230 | extracellular ligand-gated ion channel activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0060084 | synaptic transmission involved in micturition | ISS | neuron, Synap (GO term level: 8) | - |
| GO:0051963 | regulation of synaptogenesis | ISS | Synap (GO term level: 9) | - |
| GO:0021631 | optic nerve morphogenesis | ISS | Brain (GO term level: 9) | - |
| GO:0032226 | positive regulation of synaptic transmission, dopaminergic | IEA | neuron, Synap, Neurotransmitter, dopamine (GO term level: 10) | - |
| GO:0021952 | central nervous system projection neuron axonogenesis | ISS | neuron, axon (GO term level: 14) | - |
| GO:0033603 | positive regulation of dopamine secretion | ISS | dopamine (GO term level: 11) | - |
| GO:0042053 | regulation of dopamine metabolic process | ISS | dopamine (GO term level: 9) | - |
| GO:0048814 | regulation of dendrite morphogenesis | ISS | dendrite (GO term level: 13) | - |
| GO:0001666 | response to hypoxia | IDA | 12189247 | |
| GO:0001661 | conditioned taste aversion | IEA | - | |
| GO:0007165 | signal transduction | IDA | 8906617 | |
| GO:0008542 | visual learning | IMP | 15964197 | |
| GO:0007626 | locomotory behavior | ISS | - | |
| GO:0007613 | memory | IMP | 15964197 | |
| GO:0007605 | sensory perception of sound | ISS | - | |
| GO:0007601 | visual perception | ISS | - | |
| GO:0006816 | calcium ion transport | ISS | - | |
| GO:0006811 | ion transport | NAS | 8906617 | |
| GO:0006939 | smooth muscle contraction | ISS | - | |
| GO:0042220 | response to cocaine | ISS | - | |
| GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep | ISS | - | |
| GO:0021562 | vestibulocochlear nerve development | ISS | - | |
| GO:0042113 | B cell activation | ISS | - | |
| GO:0030890 | positive regulation of B cell proliferation | ISS | - | |
| GO:0019233 | sensory perception of pain | ISS | - | |
| GO:0021771 | lateral geniculate nucleus development | ISS | - | |
| GO:0035176 | social behavior | ISS | - | |
| GO:0035095 | behavioral response to nicotine | IMP | 17559419 | |
| GO:0045759 | negative regulation of action potential | ISS | - | |
| GO:0045188 | regulation of circadian sleep/wake cycle, non-REM sleep | IEA | - | |
| GO:0045471 | response to ethanol | ISS | - | |
| GO:0050890 | cognition | IMP | 17559419 | |
| GO:0051899 | membrane depolarization | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005892 | nicotinic acetylcholine-gated receptor-channel complex | IDA | Neurotransmitter (GO term level: 9) | 8906617 |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0016021 | integral to membrane | NAS | 8906617 | |
| GO:0009897 | external side of plasma membrane | ISS | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | 16 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | 13 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | 12 | 10 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |