Gene Page: COL6A2
Summary ?
| GeneID | 1292 |
| Symbol | COL6A2 |
| Synonyms | BTHLM1|PP3610|UCMD1 |
| Description | collagen type VI alpha 2 |
| Reference | MIM:120240|HGNC:HGNC:2212|Ensembl:ENSG00000142173|HPRD:02110|Vega:OTTHUMG00000090489 |
| Gene type | protein-coding |
| Map location | 21q22.3 |
| Pascal p-value | 0.019 |
| Sherlock p-value | 0.005 |
| TADA p-value | 0.02 |
| Fetal beta | 0.369 |
| eGene | Hippocampus Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| COL6A2 | chr21 | 47541029 | G | A | NM_001849 NM_058174 NM_058175 | p.484G>R p.484G>R p.484G>R | missense missense missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17131472 | chr1 | 91962966 | COL6A2 | 1292 | 0.02 | trans | ||
| rs3762382 | chr1 | 198128858 | COL6A2 | 1292 | 0.06 | trans | ||
| rs16825495 | chr2 | 134234401 | COL6A2 | 1292 | 0.09 | trans | ||
| rs7651311 | chr3 | 106820222 | COL6A2 | 1292 | 0.18 | trans | ||
| rs2922180 | chr3 | 125280472 | COL6A2 | 1292 | 0.06 | trans | ||
| rs2971271 | chr3 | 125450402 | COL6A2 | 1292 | 0.13 | trans | ||
| rs2976809 | chr3 | 125451738 | COL6A2 | 1292 | 0.01 | trans | ||
| rs313946 | chr4 | 113975904 | COL6A2 | 1292 | 0.19 | trans | ||
| rs17046025 | chr4 | 166066605 | COL6A2 | 1292 | 0.17 | trans | ||
| rs10050805 | chr5 | 30805352 | COL6A2 | 1292 | 0.07 | trans | ||
| rs317971 | chr5 | 66688724 | COL6A2 | 1292 | 0.06 | trans | ||
| rs12663450 | chr6 | 38135729 | COL6A2 | 1292 | 0.14 | trans | ||
| rs4236095 | chr6 | 47368686 | COL6A2 | 1292 | 0.1 | trans | ||
| rs6455226 | chr6 | 67930295 | COL6A2 | 1292 | 1.834E-4 | trans | ||
| rs7081666 | chr10 | 47673800 | COL6A2 | 1292 | 0.01 | trans | ||
| rs11006556 | chr10 | 61287609 | COL6A2 | 1292 | 0.07 | trans | ||
| rs10879027 | chr12 | 70237156 | COL6A2 | 1292 | 0 | trans | ||
| rs11112163 | chr12 | 105072508 | COL6A2 | 1292 | 0.06 | trans | ||
| rs6095741 | chr20 | 48666589 | COL6A2 | 1292 | 3.888E-8 | trans | ||
| rs5991662 | chrX | 43335796 | COL6A2 | 1292 | 7.397E-4 | trans | ||
| rs1545676 | chrX | 93530472 | COL6A2 | 1292 | 0.01 | trans | ||
| rs242163 | chrX | 132311662 | COL6A2 | 1292 | 0.05 | trans |
Section II. Transcriptome annotation
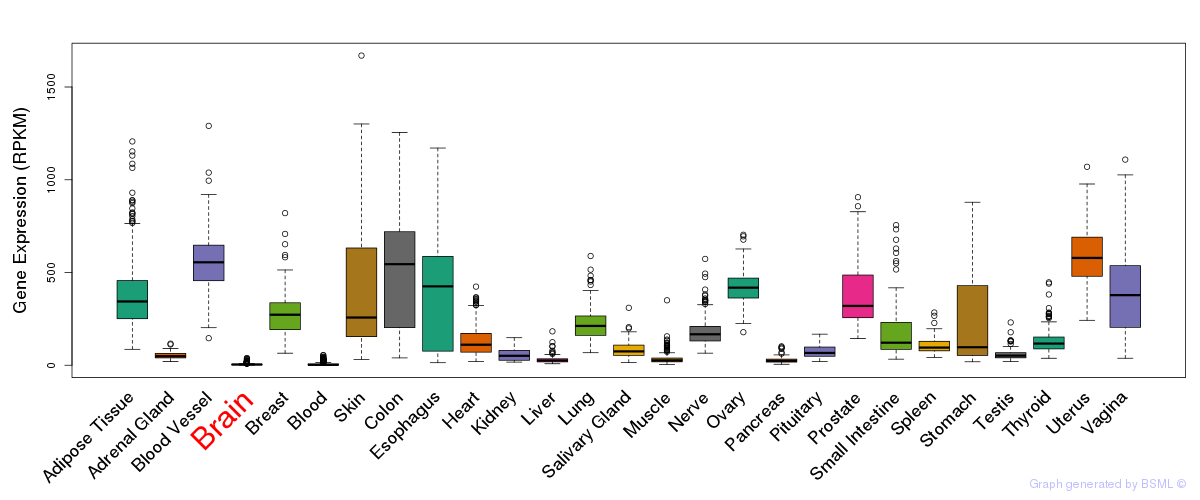
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PLCE1 | 0.89 | 0.79 |
| IQGAP2 | 0.86 | 0.66 |
| LTBP1 | 0.84 | 0.72 |
| EPHB4 | 0.84 | 0.65 |
| STON1 | 0.83 | 0.63 |
| FAM111A | 0.83 | 0.79 |
| GLI3 | 0.83 | 0.73 |
| STON1-GTF2A1L | 0.83 | 0.63 |
| KNTC1 | 0.83 | 0.58 |
| LIPG | 0.82 | 0.53 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.35 | -0.39 |
| AF347015.31 | -0.34 | -0.39 |
| C5orf53 | -0.34 | -0.34 |
| IFI27 | -0.34 | -0.37 |
| MT-CO2 | -0.34 | -0.39 |
| HLA-F | -0.34 | -0.26 |
| FBXO2 | -0.33 | -0.25 |
| CA4 | -0.33 | -0.29 |
| CISD3 | -0.33 | -0.35 |
| AF347015.33 | -0.33 | -0.34 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ECM RECEPTOR INTERACTION | 84 | 53 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN1 PATHWAY | 66 | 44 | All SZGR 2.0 genes in this pathway |
| PID AVB3 INTEGRIN PATHWAY | 75 | 53 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 1 PATHWAY | 46 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME EXTRACELLULAR MATRIX ORGANIZATION | 87 | 42 | All SZGR 2.0 genes in this pathway |
| REACTOME COLLAGEN FORMATION | 58 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME NCAM1 INTERACTIONS | 39 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | 64 | 49 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER BASAL VS MESENCHYMAL DN | 50 | 36 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F UP | 185 | 119 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION DN | 100 | 64 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| MURATA VIRULENCE OF H PILORI | 24 | 16 | All SZGR 2.0 genes in this pathway |
| WU CELL MIGRATION | 184 | 114 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 21Q22 AMPLICON | 16 | 11 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS SUBSET | 33 | 20 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION UP | 71 | 51 | All SZGR 2.0 genes in this pathway |
| POMEROY MEDULLOBLASTOMA PROGNOSIS UP | 47 | 30 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS UP | 151 | 103 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX UP | 36 | 27 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS UP | 37 | 27 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRACX UP | 20 | 14 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR DN | 91 | 64 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 4 | 48 | 32 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| JACKSON DNMT1 TARGETS DN | 25 | 21 | All SZGR 2.0 genes in this pathway |
| HAN JNK SINGALING DN | 39 | 27 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| CLASPER LYMPHATIC VESSELS DURING METASTASIS DN | 36 | 23 | All SZGR 2.0 genes in this pathway |
| HARRIS BRAIN CANCER PROGENITORS | 44 | 23 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| LEE METASTASIS AND ALTERNATIVE SPLICING UP | 74 | 51 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR DN | 191 | 123 | All SZGR 2.0 genes in this pathway |
| MISHRA CARCINOMA ASSOCIATED FIBROBLAST UP | 24 | 14 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY DN | 58 | 39 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA DN | 100 | 63 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO RADIATION THERAPY | 32 | 20 | All SZGR 2.0 genes in this pathway |
| GU PDEF TARGETS UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| SCHRAETS MLL TARGETS DN | 33 | 24 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN DEDIFFERENTIATED STATE DN | 7 | 7 | All SZGR 2.0 genes in this pathway |
| MEISSNER ES ICP WITH H3K4ME3 | 34 | 17 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC ICP WITH H3 UNMETHYLATED | 24 | 15 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN ICP WITH H3K4ME3 | 32 | 17 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 16 | 79 | 47 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN UP | 56 | 38 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA TARGETS OF HIF1A AND FOXA2 | 37 | 27 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS DN | 148 | 88 | All SZGR 2.0 genes in this pathway |
| WANG MLL TARGETS | 289 | 188 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| ANASTASSIOU CANCER MESENCHYMAL TRANSITION SIGNATURE | 64 | 40 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL MATURE DN | 99 | 74 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP | 297 | 194 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
| NABA COLLAGENS | 44 | 26 | All SZGR 2.0 genes in this pathway |
| NABA CORE MATRISOME | 275 | 148 | All SZGR 2.0 genes in this pathway |
| NABA BASEMENT MEMBRANES | 40 | 22 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |