Gene Page: CRY1
Summary ?
| GeneID | 1407 |
| Symbol | CRY1 |
| Synonyms | PHLL1 |
| Description | cryptochrome circadian clock 1 |
| Reference | MIM:601933|HGNC:HGNC:2384|Ensembl:ENSG00000008405|HPRD:09050|Vega:OTTHUMG00000170005 |
| Gene type | protein-coding |
| Map location | 12q23-q24.1 |
| Pascal p-value | 0.057 |
| Sherlock p-value | 0.048 |
| Fetal beta | 1.304 |
| eGene | Cortex Frontal Cortex BA9 Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11924592 | chr3 | 146371577 | CRY1 | 1407 | 0.09 | trans |
Section II. Transcriptome annotation
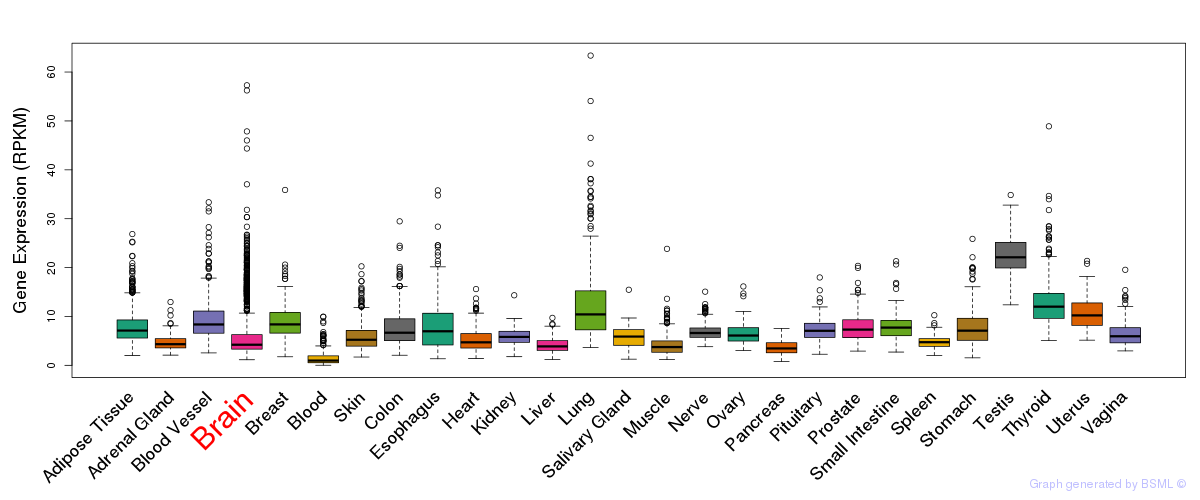
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003677 | DNA binding | TAS | 9801304 | |
| GO:0003913 | DNA photolyase activity | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0016829 | lyase activity | IEA | - | |
| GO:0008020 | G-protein coupled photoreceptor activity | TAS | 8909283 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006281 | DNA repair | IEA | - | |
| GO:0007623 | circadian rhythm | TAS | 10217146 | |
| GO:0018298 | protein-chromophore linkage | IEA | - | |
| GO:0050896 | response to stimulus | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005739 | mitochondrion | TAS | 9801304 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARNTL | BMAL1 | BMAL1c | JAP3 | MGC47515 | MOP3 | PASD3 | TIC | bHLHe5 | aryl hydrocarbon receptor nuclear translocator-like | - | HPRD,BioGRID | 10531061 |
| CBLB | DKFZp686J10223 | DKFZp779A0729 | DKFZp779F1443 | FLJ36865 | FLJ41152 | Nbla00127 | RNF56 | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | Two-hybrid | BioGRID | 16189514 |
| CRY1 | PHLL1 | cryptochrome 1 (photolyase-like) | Biochemical Activity | BioGRID | 17073458 |
| CSNK1E | HCKIE | MGC10398 | casein kinase 1, epsilon | - | HPRD | 11875063 |
| KRTAP4-12 | KAP4.12 | KRTAP4.12 | keratin associated protein 4-12 | Two-hybrid | BioGRID | 16189514 |
| MDFI | I-MF | I-mfa | MyoD family inhibitor | Two-hybrid | BioGRID | 16189514 |
| PER1 | MGC88021 | PER | RIGUI | hPER | period homolog 1 (Drosophila) | - | HPRD,BioGRID | 10531061 |
| PER2 | FASPS | KIAA0347 | period homolog 2 (Drosophila) | - | HPRD | 10428031 |10531061 |
| PER2 | FASPS | KIAA0347 | period homolog 2 (Drosophila) | Affinity Capture-Western Two-hybrid | BioGRID | 10531061 |11533252 |
| PER3 | GIG13 | period homolog 3 (Drosophila) | - | HPRD | 10428031 |
| PLSCR1 | MMTRA1B | phospholipid scramblase 1 | Two-hybrid | BioGRID | 16189514 |
| TIMELESS | FLJ12640 | FLJ20714 | TIM | TIM1 | hTIM | timeless homolog (Drosophila) | - | HPRD | 10428031 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CIRCADIAN RHYTHM MAMMAL | 13 | 13 | All SZGR 2.0 genes in this pathway |
| PID CIRCADIAN PATHWAY | 16 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | 36 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME CIRCADIAN CLOCK | 53 | 40 | All SZGR 2.0 genes in this pathway |
| SAMOLS TARGETS OF KHSV MIRNAS DN | 62 | 35 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR UP | 162 | 116 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS UP | 214 | 134 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| WIEMANN TELOMERE SHORTENING AND CHRONIC LIVER DAMAGE UP | 8 | 5 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| UEDA CENTRAL CLOCK | 88 | 62 | All SZGR 2.0 genes in this pathway |
| UEDA PERIFERAL CLOCK | 169 | 111 | All SZGR 2.0 genes in this pathway |
| MCCLUNG CREB1 TARGETS DN | 57 | 39 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 8HR UP | 105 | 73 | All SZGR 2.0 genes in this pathway |
| KANG FLUOROURACIL RESISTANCE UP | 22 | 15 | All SZGR 2.0 genes in this pathway |
| BILD E2F3 ONCOGENIC SIGNATURE | 246 | 153 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION UP | 180 | 114 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 9HR UP | 223 | 132 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| LEE RECENT THYMIC EMIGRANT | 227 | 128 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| BAE BRCA1 TARGETS UP | 75 | 47 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR UP | 199 | 143 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-361 | 312 | 318 | m8 | hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC |
| miR-381 | 184 | 190 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-448 | 624 | 631 | 1A,m8 | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-496 | 219 | 225 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.