Gene Page: MAPK14
Summary ?
| GeneID | 1432 |
| Symbol | MAPK14 |
| Synonyms | CSBP|CSBP1|CSBP2|CSPB1|EXIP|Mxi2|PRKM14|PRKM15|RK|SAPK2A|p38|p38ALPHA |
| Description | mitogen-activated protein kinase 14 |
| Reference | MIM:600289|HGNC:HGNC:6876|Ensembl:ENSG00000112062|HPRD:02619|Vega:OTTHUMG00000159806 |
| Gene type | protein-coding |
| Map location | 6p21.3-p21.2 |
| Pascal p-value | 0.699 |
| Fetal beta | 0.459 |
| eGene | Cortex |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.04433 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0305 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
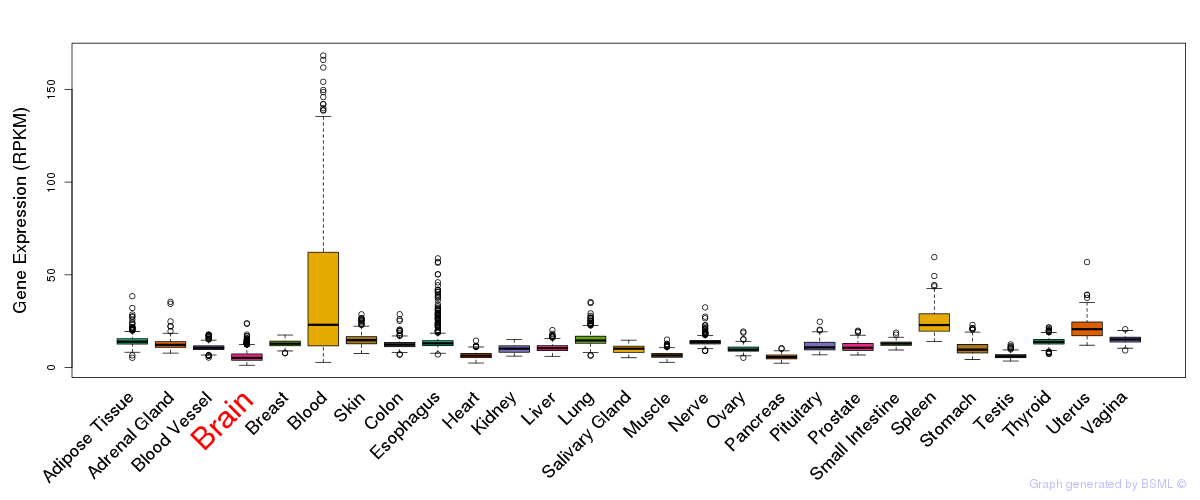
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004707 | MAP kinase activity | IEA | - | |
| GO:0004708 | MAP kinase kinase activity | TAS | 10706854 | |
| GO:0008339 | MP kinase activity | IDA | 7997261 | |
| GO:0005515 | protein binding | IPI | 9792677 |12761180 |16189514 |16636664 |16751104 |17255097 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | EXP | 8846784 |9687510 | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007265 | Ras protein signal transduction | EXP | 11520933 | |
| GO:0007243 | protein kinase cascade | IDA | 10838079 | |
| GO:0006950 | response to stress | IDA | 7997261 | |
| GO:0007166 | cell surface receptor linked signal transduction | TAS | 10706854 | |
| GO:0006935 | chemotaxis | TAS | 10706854 | |
| GO:0006928 | cell motion | TAS | 10912793 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 9687510 | |
| GO:0005634 | nucleus | ISS | - | |
| GO:0005654 | nucleoplasm | EXP | 8846784 | |
| GO:0005737 | cytoplasm | ISS | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | Affinity Capture-Western Biochemical Activity | BioGRID | 11042204 |
| ATF2 | CRE-BP1 | CREB2 | HB16 | MGC111558 | TREB7 | activating transcription factor 2 | An unspecified isoform of p38 interacts with and phosphorylates ATF2. | BIND | 9155018 |
| ATF2 | CRE-BP1 | CREB2 | HB16 | MGC111558 | TREB7 | activating transcription factor 2 | An unspecified isoform of p38 interacts with and phosphorylates ATF2. | BIND | 9235954 |
| ATF2 | CRE-BP1 | CREB2 | HB16 | MGC111558 | TREB7 | activating transcription factor 2 | p38 phosphorylates ATF2. This interaction was modelled on a demonstrated interaction between human p38 and ATF2 from an unspecified species. | BIND | 15692053 |
| ATF2 | CRE-BP1 | CREB2 | HB16 | MGC111558 | TREB7 | activating transcription factor 2 | - | HPRD,BioGRID | 7535770 |
| CDC25A | CDC25A2 | cell division cycle 25 homolog A (S. pombe) | Reconstituted Complex | BioGRID | 12963847 |
| CDC25B | - | cell division cycle 25 homolog B (S. pombe) | Biochemical Activity Reconstituted Complex | BioGRID | 11333986 |
| CDC25B | - | cell division cycle 25 homolog B (S. pombe) | p38-alpha interacts with and phosphorylates CDC25B. | BIND | 15629715 |
| CDC25C | CDC25 | cell division cycle 25 homolog C (S. pombe) | - | HPRD,BioGRID | 11333986 |
| CREB1 | CREB | MGC9284 | cAMP responsive element binding protein 1 | - | HPRD,BioGRID | 11377386 |
| CSNK2A1 | CK2A1 | CKII | casein kinase 2, alpha 1 polypeptide | Affinity Capture-Western Reconstituted Complex | BioGRID | 10747897 |
| CSNK2A2 | CK2A2 | CSNK2A1 | FLJ43934 | casein kinase 2, alpha prime polypeptide | - | HPRD | 10747897 |
| CSNK2B | CK2B | CK2N | CSK2B | G5A | MGC138222 | MGC138224 | casein kinase 2, beta polypeptide | - | HPRD | 10747897 |
| DDIT3 | CEBPZ | CHOP | CHOP10 | GADD153 | MGC4154 | DNA-damage-inducible transcript 3 | - | HPRD,BioGRID | 8650547 |
| DUSP1 | CL100 | HVH1 | MKP-1 | MKP1 | PTPN10 | dual specificity phosphatase 1 | - | HPRD | 7535770 |
| DUSP1 | CL100 | HVH1 | MKP-1 | MKP1 | PTPN10 | dual specificity phosphatase 1 | Affinity Capture-Western Two-hybrid | BioGRID | 11278799 |11359773 |
| DUSP10 | MKP-5 | MKP5 | dual specificity phosphatase 10 | Affinity Capture-Western Biochemical Activity Reconstituted Complex Two-hybrid | BioGRID | 10391943 |11157753 |11359773 |
| DUSP10 | MKP-5 | MKP5 | dual specificity phosphatase 10 | An unspecified isoform of MAPK14 interacts with DUSP10. | BIND | 10391943 |
| DUSP10 | MKP-5 | MKP5 | dual specificity phosphatase 10 | - | HPRD | 10391943 |10597297 |11359773 |
| DUSP16 | KIAA1700 | MGC129701 | MGC129702 | MKP-7 | MKP7 | dual specificity phosphatase 16 | - | HPRD,BioGRID | 11359773 |11489891 |14586399 |
| DUSP2 | PAC-1 | PAC1 | dual specificity phosphatase 2 | - | HPRD | 7535770 |
| DUSP22 | JKAP | JSP1 | MKPX | VHX | dual specificity phosphatase 22 | - | HPRD | 11346645 |
| DUSP4 | HVH2 | MKP-2 | MKP2 | TYP | dual specificity phosphatase 4 | - | HPRD,BioGRID | 11387337 |
| DUSP6 | MKP3 | PYST1 | dual specificity phosphatase 6 | Affinity Capture-Western | BioGRID | 11359773 |
| DYRK1B | MIRK | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B | - | HPRD,BioGRID | 10910078 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | Biochemical Activity | BioGRID | 7535770 |
| ELK1 | - | ELK1, member of ETS oncogene family | Elk-1 is phosphorylated by an unspecified isoform of p38 MAPK. This interaction was modeled on a demonstrated interaction between human Elk-1 and p38 MAPK from an unspecified species. | BIND | 9130707 |
| ELK1 | - | ELK1, member of ETS oncogene family | - | HPRD | 8622669 |
| ELK1 | - | ELK1, member of ETS oncogene family | An unspecified isoform of p38 interacts with and phosphorylates Elk-1. | BIND | 9155018 |
| ELK4 | SAP1 | ELK4, ETS-domain protein (SRF accessory protein 1) | An unspecified isoform of p38 interacts with and phosphorylates Sap-1a. | BIND | 9235954 |
| ELK4 | SAP1 | ELK4, ETS-domain protein (SRF accessory protein 1) | Sap-1a is phosphorylated by an unspecified isoform of p38 MAPK. This interaction was modeled on a demonstrated interaction between human Sap-1a and p38 MAPK from an unspecified species. | BIND | 9130707 |
| ETV1 | DKFZp781L0674 | ER81 | MGC104699 | MGC120533 | MGC120534 | ets variant 1 | Biochemical Activity | BioGRID | 11551945 |
| FLNA | ABP-280 | ABPX | DKFZp434P031 | FLN | FLN1 | FMD | MNS | NHBP | OPD | OPD1 | OPD2 | filamin A, alpha (actin binding protein 280) | - | HPRD | 11390380 |
| FUBP1 | FBP | FUBP | far upstream element (FUSE) binding protein 1 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12819782 |
| GMFB | GMF | glia maturation factor, beta | - | HPRD,BioGRID | 8798479 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD | 8695800 |
| HTRA2 | OMI | PARK13 | PRSS25 | HtrA serine peptidase 2 | - | HPRD,BioGRID | 10644717 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | An unspecified isoform of p38 interacts with and phosphorylates c-Jun. | BIND | 9155018 |
| KARS | KARS2 | KIAA0070 | KRS | lysyl-tRNA synthetase | Two-hybrid | BioGRID | 12819782 |
| KRT18 | CYK18 | K18 | keratin 18 | - | HPRD,BioGRID | 11788583 |
| MAP2K1 | MAPKK1 | MEK1 | MKK1 | PRKMK1 | mitogen-activated protein kinase kinase 1 | - | HPRD | 12697810 |
| MAP2K3 | MAPKK3 | MEK3 | MKK3 | PRKMK3 | mitogen-activated protein kinase kinase 3 | - | HPRD | 7839144|8622669 |
| MAP2K3 | MAPKK3 | MEK3 | MKK3 | PRKMK3 | mitogen-activated protein kinase kinase 3 | Biochemical Activity | BioGRID | 11279118 |
| MAP2K3 | MAPKK3 | MEK3 | MKK3 | PRKMK3 | mitogen-activated protein kinase kinase 3 | MKK3 phosphorylates an unspecified isoform of p38. This interaction was modeled on a demonstrated interaction between human MKK3 and p38 from an unspecified species. | BIND | 8622669 |
| MAP2K4 | JNKK | JNKK1 | MAPKK4 | MEK4 | MKK4 | PRKMK4 | SEK1 | SERK1 | mitogen-activated protein kinase kinase 4 | - | HPRD,BioGRID | 7716521 |
| MAP2K6 | MAPKK6 | MEK6 | MKK6 | PRKMK6 | SAPKK3 | mitogen-activated protein kinase kinase 6 | - | HPRD,BioGRID | 8626699 |
| MAP2K7 | Jnkk2 | MAPKK7 | MKK7 | PRKMK7 | mitogen-activated protein kinase kinase 7 | Biochemical Activity | BioGRID | 9207092 |
| MAP3K7IP1 | 3'-Tab1 | MGC57664 | TAB1 | mitogen-activated protein kinase kinase kinase 7 interacting protein 1 | TAB1 interacts with an unspecified isoform of p38. | BIND | 12829618 |
| MAP3K7IP1 | 3'-Tab1 | MGC57664 | TAB1 | mitogen-activated protein kinase kinase kinase 7 interacting protein 1 | - | HPRD,BioGRID | 11847341 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | - | HPRD,BioGRID | 12697810 |
| MAPK14 | CSBP1 | CSBP2 | CSPB1 | EXIP | Mxi2 | PRKM14 | PRKM15 | RK | SAPK2A | p38 | p38ALPHA | mitogen-activated protein kinase 14 | Biochemical Activity Two-hybrid | BioGRID | 11847341 |12819782 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | - | HPRD,BioGRID | 12697810 |
| MAPKAPK2 | MK2 | mitogen-activated protein kinase-activated protein kinase 2 | - | HPRD,BioGRID | 9768359 |11042204 |11551945 |
| MAPKAPK3 | 3PK | MAPKAP3 | mitogen-activated protein kinase-activated protein kinase 3 | - | HPRD,BioGRID | 11157753 |
| MARS | FLJ35667 | METRS | MTRNS | methionyl-tRNA synthetase | - | HPRD | 9878398 |
| MEF2A | ADCAD1 | RSRFC4 | RSRFC9 | myocyte enhancer factor 2A | - | HPRD,BioGRID | 10330143 |
| MEF2C | - | myocyte enhancer factor 2C | - | HPRD | 9753748 |10330143 |
| MKNK1 | MNK1 | MAP kinase interacting serine/threonine kinase 1 | - | HPRD | 9155018 |
| MKNK1 | MNK1 | MAP kinase interacting serine/threonine kinase 1 | Affinity Capture-Western | BioGRID | 11157753 |
| MKNK1 | MNK1 | MAP kinase interacting serine/threonine kinase 1 | An unspecified isoform of p38 interacts with and phosphorylates MNK1. | BIND | 9155018 |
| MKNK2 | GPRK7 | MNK2 | MAP kinase interacting serine/threonine kinase 2 | - | HPRD | 12897141 |
| NCF1C | SH3PXD1C | neutrophil cytosolic factor 1C pseudogene | p38-alpha interacts with and phosphorylates p47phox. | BIND | 15629715 |
| PLCB2 | FLJ38135 | phospholipase C, beta 2 | Affinity Capture-Western | BioGRID | 12054652 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | - | HPRD | 15273249 |
| PTPN7 | BPTP-4 | HEPTP | LC-PTP | LPTP | PTPNI | protein tyrosine phosphatase, non-receptor type 7 | - | HPRD | 10559944 |
| RPS6KA3 | CLS | HU-3 | ISPK-1 | MAPKAPK1B | MRX19 | RSK | RSK2 | S6K-alpha3 | p90-RSK2 | pp90RSK2 | ribosomal protein S6 kinase, 90kDa, polypeptide 3 | Affinity Capture-Western | BioGRID | 11157753 |
| RPS6KA4 | MSK2 | RSK-B | ribosomal protein S6 kinase, 90kDa, polypeptide 4 | - | HPRD,BioGRID | 9792677 |
| RPS6KA5 | MGC1911 | MSK1 | MSPK1 | RLPK | ribosomal protein S6 kinase, 90kDa, polypeptide 5 | Affinity Capture-Western | BioGRID | 11157753 |
| SCYE1 | AIMP1 | EMAP-2 | EMAP-II | EMAP2 | EMAPII | p43 | small inducible cytokine subfamily E, member 1 (endothelial monocyte-activating) | - | HPRD | 9878398 |
| SLC12A2 | BSC | BSC2 | MGC104233 | NKCC1 | solute carrier family 12 (sodium/potassium/chloride transporters), member 2 | Affinity Capture-Western | BioGRID | 14563843 |
| SLC44A1 | CD92 | CDW92 | CHTL1 | CTL1 | RP11-287A8.1 | solute carrier family 44, member 1 | - | HPRD | 9707433 |
| SMAD7 | CRCS3 | FLJ16482 | MADH7 | MADH8 | SMAD family member 7 | - | HPRD,BioGRID | 12589052 |
| SPAG9 | FLJ13450 | FLJ14006 | FLJ26141 | FLJ34602 | HLC4 | JLP | KIAA0516 | MGC117291 | MGC14967 | MGC74461 | PHET | PIG6 | sperm associated antigen 9 | - | HPRD,BioGRID | 12391307 |
| STK39 | DCHT | DKFZp686K05124 | PASK | SPAK | serine threonine kinase 39 (STE20/SPS1 homolog, yeast) | - | HPRD | 10980603 |
| STK39 | DCHT | DKFZp686K05124 | PASK | SPAK | serine threonine kinase 39 (STE20/SPS1 homolog, yeast) | Affinity Capture-Western | BioGRID | 14563843 |
| ZFP36L1 | BRF1 | Berg36 | ERF-1 | ERF1 | RNF162B | TIS11B | cMG1 | zinc finger protein 36, C3H type-like 1 | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG VEGF SIGNALING PATHWAY | 76 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| KEGG NOD LIKE RECEPTOR SIGNALING PATHWAY | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG RIG I LIKE RECEPTOR SIGNALING PATHWAY | 71 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| KEGG FC EPSILON RI SIGNALING PATHWAY | 79 | 58 | All SZGR 2.0 genes in this pathway |
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG GNRH SIGNALING PATHWAY | 101 | 77 | All SZGR 2.0 genes in this pathway |
| KEGG PROGESTERONE MEDIATED OOCYTE MATURATION | 86 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG AMYOTROPHIC LATERAL SCLEROSIS ALS | 53 | 43 | All SZGR 2.0 genes in this pathway |
| KEGG EPITHELIAL CELL SIGNALING IN HELICOBACTER PYLORI INFECTION | 68 | 44 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION | 72 | 56 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BCR PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BIOPEPTIDES PATHWAY | 45 | 35 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HDAC PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FMLP PATHWAY | 39 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GATA3 PATHWAY | 16 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HCMV PATHWAY | 17 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL12 PATHWAY | 23 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA KERATINOCYTE PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PYK2 PATHWAY | 31 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EGFR SMRTE PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NFAT PATHWAY | 56 | 45 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NTHI PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ARENRF2 PATHWAY | 13 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P38MAPK PATHWAY | 40 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CCR5 PATHWAY | 20 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RB PATHWAY | 13 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EIF4 PATHWAY | 24 | 19 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL1R PATHWAY | 33 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TALL1 PATHWAY | 15 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA 41BB PATHWAY | 17 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SARS PATHWAY | 10 | 5 | All SZGR 2.0 genes in this pathway |
| BIOCARTA STRESS PATHWAY | 25 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TOLL PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CREB PATHWAY | 27 | 25 | All SZGR 2.0 genes in this pathway |
| SIG CD40PATHWAYMAP | 34 | 28 | All SZGR 2.0 genes in this pathway |
| ST P38 MAPK PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| ST ADRENERGIC | 36 | 29 | All SZGR 2.0 genes in this pathway |
| PID ENDOTHELIN PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| PID BCR 5PATHWAY | 65 | 50 | All SZGR 2.0 genes in this pathway |
| PID P73PATHWAY | 79 | 59 | All SZGR 2.0 genes in this pathway |
| PID P38 MKK3 6PATHWAY | 26 | 21 | All SZGR 2.0 genes in this pathway |
| PID NFKAPPAB ATYPICAL PATHWAY | 17 | 15 | All SZGR 2.0 genes in this pathway |
| PID IL4 2PATHWAY | 65 | 43 | All SZGR 2.0 genes in this pathway |
| PID CD40 PATHWAY | 31 | 26 | All SZGR 2.0 genes in this pathway |
| PID IL12 2PATHWAY | 63 | 54 | All SZGR 2.0 genes in this pathway |
| PID P38 ALPHA BETA PATHWAY | 31 | 25 | All SZGR 2.0 genes in this pathway |
| PID CDC42 PATHWAY | 70 | 51 | All SZGR 2.0 genes in this pathway |
| PID ANGIOPOIETIN RECEPTOR PATHWAY | 50 | 41 | All SZGR 2.0 genes in this pathway |
| PID FAS PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| PID TXA2PATHWAY | 57 | 43 | All SZGR 2.0 genes in this pathway |
| PID NFAT 3PATHWAY | 54 | 47 | All SZGR 2.0 genes in this pathway |
| PID REG GR PATHWAY | 82 | 60 | All SZGR 2.0 genes in this pathway |
| PID IL2 1PATHWAY | 55 | 43 | All SZGR 2.0 genes in this pathway |
| PID AR TF PATHWAY | 53 | 38 | All SZGR 2.0 genes in this pathway |
| PID ATF2 PATHWAY | 59 | 43 | All SZGR 2.0 genes in this pathway |
| PID IL6 7 PATHWAY | 47 | 40 | All SZGR 2.0 genes in this pathway |
| PID P38 MK2 PATHWAY | 21 | 19 | All SZGR 2.0 genes in this pathway |
| PID RETINOIC ACID PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| PID CXCR3 PATHWAY | 43 | 34 | All SZGR 2.0 genes in this pathway |
| PID P38 ALPHA BETA DOWNSTREAM PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| PID EPO PATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PID VEGFR1 2 PATHWAY | 69 | 57 | All SZGR 2.0 genes in this pathway |
| PID P53 REGULATION PATHWAY | 59 | 50 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P2 PATHWAY | 24 | 19 | All SZGR 2.0 genes in this pathway |
| PID MAPK TRK PATHWAY | 34 | 31 | All SZGR 2.0 genes in this pathway |
| PID LYMPH ANGIOGENESIS PATHWAY | 25 | 16 | All SZGR 2.0 genes in this pathway |
| PID RAC1 PATHWAY | 54 | 37 | All SZGR 2.0 genes in this pathway |
| PID RB 1PATHWAY | 65 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME TRIF MEDIATED TLR3 SIGNALING | 74 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING TO RAS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING TO ERKS | 36 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME P38MAPK EVENTS | 13 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | 24 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME ERK MAPK TARGETS | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME ADP SIGNALLING THROUGH P2RY1 | 25 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNAL AMPLIFICATION | 31 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME MYOGENESIS | 28 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME MAP KINASE ACTIVATION IN TLR CASCADE | 50 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | 10 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME MAPK TARGETS NUCLEAR EVENTS MEDIATED BY MAP KINASES | 30 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | 18 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET HOMEOSTASIS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET SENSITIZATION BY LDL | 16 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | 84 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | 77 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME DESTABILIZATION OF MRNA BY KSRP | 17 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB AND MAP KINASES ACTIVATION MEDIATED BY TLR4 SIGNALING REPERTOIRE | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME DSCAM INTERACTIONS | 11 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING | 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME NOD1 2 SIGNALING PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | 46 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN | 384 | 220 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 UP | 64 | 46 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR DN | 64 | 39 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR DN | 75 | 50 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK DN | 137 | 97 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP | 197 | 135 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS UP | 169 | 127 | All SZGR 2.0 genes in this pathway |
| GALLUZZI PREVENT MITOCHONDIAL PERMEABILIZATION | 22 | 16 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO BEXAROTENE DN | 29 | 19 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 ONLY DN | 44 | 30 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| RANKIN ANGIOGENIC TARGETS OF VHL HIF2A UP | 5 | 5 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR DN | 41 | 30 | All SZGR 2.0 genes in this pathway |
| UEDA PERIFERAL CLOCK | 169 | 111 | All SZGR 2.0 genes in this pathway |
| TENEDINI MEGAKARYOCYTE MARKERS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT REJECTED VS OK DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| PARK HSC AND MULTIPOTENT PROGENITORS | 50 | 33 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| HOEGERKORP CD44 TARGETS DIRECT UP | 27 | 21 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 6HR UP | 37 | 31 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE DN | 123 | 76 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 8 HR DN | 53 | 40 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D8 | 40 | 29 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS DN | 87 | 66 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN UP | 90 | 58 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE UP | 242 | 159 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 605 | 612 | 1A,m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 604 | 611 | 1A,m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-125/351 | 1610 | 1616 | m8 | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-128 | 32 | 39 | 1A,m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-142-5p | 1589 | 1596 | 1A,m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-185 | 62 | 68 | 1A | hsa-miR-185brain | UGGAGAGAAAGGCAGUUC |
| miR-19 | 2057 | 2064 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-22 | 1451 | 1457 | m8 | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-224 | 213 | 219 | m8 | hsa-miR-224 | CAAGUCACUAGUGGUUCCGUUUA |
| miR-27 | 33 | 39 | m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-505 | 1105 | 1112 | 1A,m8 | hsa-miR-505 | GUCAACACUUGCUGGUUUCCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.