Gene Page: CTSB
Summary ?
| GeneID | 1508 |
| Symbol | CTSB |
| Synonyms | APPS|CPSB |
| Description | cathepsin B |
| Reference | MIM:116810|HGNC:HGNC:2527|Ensembl:ENSG00000164733|HPRD:00287|Vega:OTTHUMG00000090799 |
| Gene type | protein-coding |
| Map location | 8p22 |
| Pascal p-value | 0.007 |
| Sherlock p-value | 0.697 |
| DMG | 2 (# studies) |
| eGene | Caudate basal ganglia Cerebellum Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1211 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02637136 | 8 | 11725599 | CTSB | 1.704E-4 | -0.349 | 0.033 | DMG:Wockner_2014 |
| cg18787975 | 8 | 11726094 | CTSB | 8.74E-8 | -0.012 | 2.01E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| snp_a-1856993 | 0 | CTSB | 1508 | 0.2 | trans | |||
| rs35558975 | 8 | 11584476 | CTSB | ENSG00000164733.16 | 1.123E-6 | 0 | 142481 | gtex_brain_putamen_basal |
| rs1736065 | 8 | 11685453 | CTSB | ENSG00000164733.16 | 5.745E-7 | 0 | 41504 | gtex_brain_putamen_basal |
| rs144275726 | 8 | 11685801 | CTSB | ENSG00000164733.16 | 1.2E-6 | 0 | 41156 | gtex_brain_putamen_basal |
| rs1293316 | 8 | 11690915 | CTSB | ENSG00000164733.16 | 5.838E-7 | 0 | 36042 | gtex_brain_putamen_basal |
| rs6990033 | 8 | 11693378 | CTSB | ENSG00000164733.16 | 2.796E-7 | 0 | 33579 | gtex_brain_putamen_basal |
| rs1293325 | 8 | 11693577 | CTSB | ENSG00000164733.16 | 5.838E-7 | 0 | 33380 | gtex_brain_putamen_basal |
| rs1293326 | 8 | 11694765 | CTSB | ENSG00000164733.16 | 5.838E-7 | 0 | 32192 | gtex_brain_putamen_basal |
| rs1293327 | 8 | 11694986 | CTSB | ENSG00000164733.16 | 4.256E-7 | 0 | 31971 | gtex_brain_putamen_basal |
| rs1293328 | 8 | 11695060 | CTSB | ENSG00000164733.16 | 5.838E-7 | 0 | 31897 | gtex_brain_putamen_basal |
| rs1296026 | 8 | 11697442 | CTSB | ENSG00000164733.16 | 5.025E-7 | 0 | 29515 | gtex_brain_putamen_basal |
| rs1293331 | 8 | 11698436 | CTSB | ENSG00000164733.16 | 5.102E-7 | 0 | 28521 | gtex_brain_putamen_basal |
| rs1296028 | 8 | 11698747 | CTSB | ENSG00000164733.16 | 5.838E-7 | 0 | 28210 | gtex_brain_putamen_basal |
| rs1613883 | 8 | 11699632 | CTSB | ENSG00000164733.16 | 5.756E-7 | 0 | 27325 | gtex_brain_putamen_basal |
| rs1692821 | 8 | 11699988 | CTSB | ENSG00000164733.16 | 7.886E-7 | 0 | 26969 | gtex_brain_putamen_basal |
| rs2740592 | 8 | 11701253 | CTSB | ENSG00000164733.16 | 5.102E-7 | 0 | 25704 | gtex_brain_putamen_basal |
| rs2645425 | 8 | 11701278 | CTSB | ENSG00000164733.16 | 5.114E-7 | 0 | 25679 | gtex_brain_putamen_basal |
| rs4839 | 8 | 11701933 | CTSB | ENSG00000164733.16 | 5.102E-7 | 0 | 25024 | gtex_brain_putamen_basal |
| rs3947 | 8 | 11702375 | CTSB | ENSG00000164733.16 | 5.154E-7 | 0 | 24582 | gtex_brain_putamen_basal |
| rs709821 | 8 | 11702594 | CTSB | ENSG00000164733.16 | 5.197E-7 | 0 | 24363 | gtex_brain_putamen_basal |
| rs1736081 | 8 | 11702840 | CTSB | ENSG00000164733.16 | 2.995E-7 | 0 | 24117 | gtex_brain_putamen_basal |
| rs1692811 | 8 | 11703048 | CTSB | ENSG00000164733.16 | 5.34E-7 | 0 | 23909 | gtex_brain_putamen_basal |
| rs1736082 | 8 | 11703420 | CTSB | ENSG00000164733.16 | 5.102E-7 | 0 | 23537 | gtex_brain_putamen_basal |
| rs1736083 | 8 | 11703497 | CTSB | ENSG00000164733.16 | 5.544E-7 | 0 | 23460 | gtex_brain_putamen_basal |
| rs1692812 | 8 | 11703523 | CTSB | ENSG00000164733.16 | 5.554E-7 | 0 | 23434 | gtex_brain_putamen_basal |
| rs1736084 | 8 | 11703657 | CTSB | ENSG00000164733.16 | 5.102E-7 | 0 | 23300 | gtex_brain_putamen_basal |
| rs1736085 | 8 | 11703659 | CTSB | ENSG00000164733.16 | 5.102E-7 | 0 | 23298 | gtex_brain_putamen_basal |
| rs1736086 | 8 | 11704897 | CTSB | ENSG00000164733.16 | 1.766E-6 | 0 | 22060 | gtex_brain_putamen_basal |
| rs1692819 | 8 | 11705448 | CTSB | ENSG00000164733.16 | 9.144E-8 | 0 | 21509 | gtex_brain_putamen_basal |
| rs1736089 | 8 | 11705870 | CTSB | ENSG00000164733.16 | 6.551E-8 | 0 | 21087 | gtex_brain_putamen_basal |
| rs2740594 | 8 | 11707174 | CTSB | ENSG00000164733.16 | 5.048E-8 | 0 | 19783 | gtex_brain_putamen_basal |
| rs2645417 | 8 | 11708800 | CTSB | ENSG00000164733.16 | 9.882E-8 | 0 | 18157 | gtex_brain_putamen_basal |
| rs2740595 | 8 | 11708830 | CTSB | ENSG00000164733.16 | 4.785E-8 | 0 | 18127 | gtex_brain_putamen_basal |
| rs1293289 | 8 | 11709945 | CTSB | ENSG00000164733.16 | 2.317E-6 | 0 | 17012 | gtex_brain_putamen_basal |
| rs1293292 | 8 | 11710369 | CTSB | ENSG00000164733.16 | 2.241E-6 | 0 | 16588 | gtex_brain_putamen_basal |
| rs1122182 | 8 | 11710629 | CTSB | ENSG00000164733.16 | 2.195E-6 | 0 | 16328 | gtex_brain_putamen_basal |
| rs34504234 | 8 | 11711113 | CTSB | ENSG00000164733.16 | 6.443E-7 | 0 | 15844 | gtex_brain_putamen_basal |
| rs1293296 | 8 | 11711157 | CTSB | ENSG00000164733.16 | 6.474E-7 | 0 | 15800 | gtex_brain_putamen_basal |
| rs1293298 | 8 | 11712443 | CTSB | ENSG00000164733.16 | 5.409E-7 | 0 | 14514 | gtex_brain_putamen_basal |
| rs1736103 | 8 | 11712639 | CTSB | ENSG00000164733.16 | 3.941E-8 | 0 | 14318 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
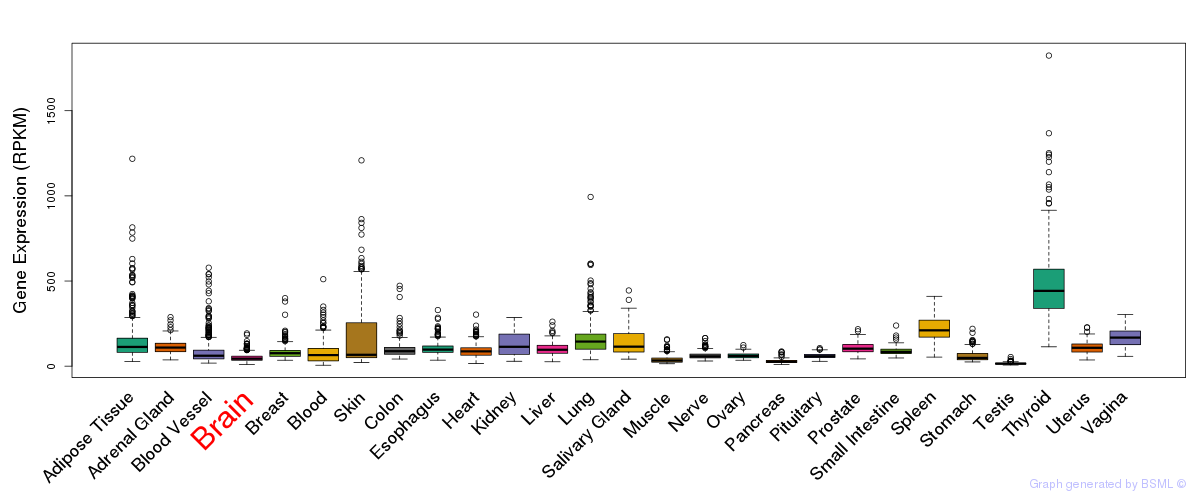
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 16364318 | |
| GO:0004197 | cysteine-type endopeptidase activity | IEA | - | |
| GO:0004197 | cysteine-type endopeptidase activity | TAS | 1645961 | |
| GO:0008233 | peptidase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006508 | proteolysis | IEA | - | |
| GO:0006508 | proteolysis | TAS | 3463996 | |
| GO:0042981 | regulation of apoptosis | TAS | 16130169 | |
| GO:0050790 | regulation of catalytic activity | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | TAS | 1645961 | |
| GO:0005739 | mitochondrion | IEA | - | |
| GO:0005764 | lysosome | IEA | - | |
| GO:0042470 | melanosome | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| A2M | CPAMD5 | DKFZp779B086 | FWP007 | S863-7 | alpha-2-macroglobulin | - | HPRD | 2476070 |
| ANXA2 | ANX2 | ANX2L4 | CAL1H | LIP2 | LPC2 | LPC2D | P36 | PAP-IV | annexin A2 | - | HPRD,BioGRID | 10777578 |
| APOE | AD2 | LPG | MGC1571 | apoprotein | apolipoprotein E | - | HPRD | 12057992 |
| BAT3 | BAG-6 | BAG6 | D6S52E | G3 | HLA-B associated transcript 3 | Two-hybrid | BioGRID | 16169070 |
| CST3 | ARMD11 | MGC117328 | cystatin C | Reconstituted Complex | BioGRID | 14503883 |
| CST3 | ARMD11 | MGC117328 | cystatin C | - | HPRD | 1996959 |7890620 |
| CST7 | CMAP | cystatin F (leukocystatin) | - | HPRD | 9632704 |9733783 |
| CSTA | STF1 | STFA | cystatin A (stefin A) | Reconstituted Complex | BioGRID | 9585570 |14503883 |
| CSTA | STF1 | STFA | cystatin A (stefin A) | - | HPRD | 9585570 |10353845 |14503883 |
| CSTB | CST6 | EPM1 | PME | STFB | cystatin B (stefin B) | - | HPRD,BioGRID | 11514663 |
| MARCKS | 80K-L | FLJ14368 | FLJ90045 | MACS | MRACKS | PKCSL | PRKCSL | myristoylated alanine-rich protein kinase C substrate | - | HPRD | 9295331 |
| S100A10 | 42C | ANX2L | ANX2LG | CAL1L | CLP11 | Ca[1] | GP11 | MGC111133 | P11 | p10 | S100 calcium binding protein A10 | - | HPRD,BioGRID | 10777578 |
| SERPINB13 | HSHUR7SEQ | HUR7 | MGC126870 | PI13 | headpin | serpin peptidase inhibitor, clade B (ovalbumin), member 13 | - | HPRD | 10512713 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG LYSOSOME | 121 | 83 | All SZGR 2.0 genes in this pathway |
| KEGG ANTIGEN PROCESSING AND PRESENTATION | 89 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME MHC CLASS II ANTIGEN PRESENTATION | 91 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS DN | 186 | 114 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| SMIRNOV CIRCULATING ENDOTHELIOCYTES IN CANCER UP | 158 | 103 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D DN | 205 | 127 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D DN | 142 | 90 | All SZGR 2.0 genes in this pathway |
| PAPASPYRIDONOS UNSTABLE ATEROSCLEROTIC PLAQUE UP | 52 | 36 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL DN | 226 | 132 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS DN | 136 | 94 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS UP | 126 | 72 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 UP | 120 | 73 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF UP | 215 | 137 | All SZGR 2.0 genes in this pathway |
| CHIN BREAST CANCER COPY NUMBER DN | 16 | 10 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| MISSIAGLIA REGULATED BY METHYLATION UP | 126 | 78 | All SZGR 2.0 genes in this pathway |
| WANG BARRETTS ESOPHAGUS DN | 25 | 13 | All SZGR 2.0 genes in this pathway |
| AIYAR COBRA1 TARGETS UP | 39 | 25 | All SZGR 2.0 genes in this pathway |
| DAUER STAT3 TARGETS UP | 49 | 35 | All SZGR 2.0 genes in this pathway |
| TSUNODA CISPLATIN RESISTANCE DN | 51 | 38 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| HOWLIN PUBERTAL MAMMARY GLAND | 69 | 40 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| LIU BREAST CANCER | 30 | 19 | All SZGR 2.0 genes in this pathway |
| WEI MIR34A TARGETS | 148 | 97 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| MORI PLASMA CELL UP | 51 | 29 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION UP | 71 | 51 | All SZGR 2.0 genes in this pathway |
| WIELAND UP BY HBV INFECTION | 101 | 66 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGLL VS IGLK UP | 42 | 24 | All SZGR 2.0 genes in this pathway |
| NADLER OBESITY UP | 61 | 34 | All SZGR 2.0 genes in this pathway |
| LIAN NEUTROPHIL GRANULE CONSTITUENTS | 25 | 13 | All SZGR 2.0 genes in this pathway |
| APPEL IMATINIB RESPONSE | 33 | 22 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS MATURE CELL | 293 | 160 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS DN | 74 | 55 | All SZGR 2.0 genes in this pathway |
| LIAN LIPA TARGETS 6M | 74 | 47 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS UP | 56 | 35 | All SZGR 2.0 genes in this pathway |
| CHEN LVAD SUPPORT OF FAILING HEART DN | 42 | 32 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| LIAN LIPA TARGETS 3M | 59 | 36 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES DN | 245 | 144 | All SZGR 2.0 genes in this pathway |
| KYNG RESPONSE TO H2O2 | 71 | 42 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES UP | 253 | 147 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND MACROPHAGE | 77 | 50 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND FIBROBLAST | 132 | 93 | All SZGR 2.0 genes in this pathway |
| KONDO EZH2 TARGETS | 245 | 148 | All SZGR 2.0 genes in this pathway |
| ZHANG GATA6 TARGETS DN | 64 | 46 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN UP | 181 | 112 | All SZGR 2.0 genes in this pathway |
| OUYANG PROSTATE CANCER MARKERS | 19 | 16 | All SZGR 2.0 genes in this pathway |
| HUANG FOXA2 TARGETS DN | 36 | 21 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| CHUNG BLISTER CYTOTOXICITY UP | 134 | 84 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS EMT UP | 36 | 24 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| HELLEBREKERS SILENCED DURING TUMOR ANGIOGENESIS | 80 | 56 | All SZGR 2.0 genes in this pathway |
| TOOKER GEMCITABINE RESISTANCE DN | 122 | 84 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G1 DN | 40 | 26 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS DN | 207 | 139 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA MESENCHYMAL | 216 | 130 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE UP | 242 | 159 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |
| GREGORY SYNTHETIC LETHAL WITH IMATINIB | 145 | 83 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 1 DN | 63 | 39 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| DELPUECH FOXO3 TARGETS UP | 68 | 49 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| NABA ECM REGULATORS | 238 | 125 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-186 | 59 | 66 | 1A,m8 | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.