Gene Page: DCC
Summary ?
| GeneID | 1630 |
| Symbol | DCC |
| Synonyms | CRC18|CRCR1|IGDCC1|MRMV1|NTN1R1 |
| Description | DCC netrin 1 receptor |
| Reference | MIM:120470|HGNC:HGNC:2701|Ensembl:ENSG00000187323|HPRD:00391|Vega:OTTHUMG00000132698 |
| Gene type | protein-coding |
| Map location | 18q21.3 |
| Fetal beta | 2.066 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23242898 | 18 | 49867273 | DCC | -0.023 | 0.29 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
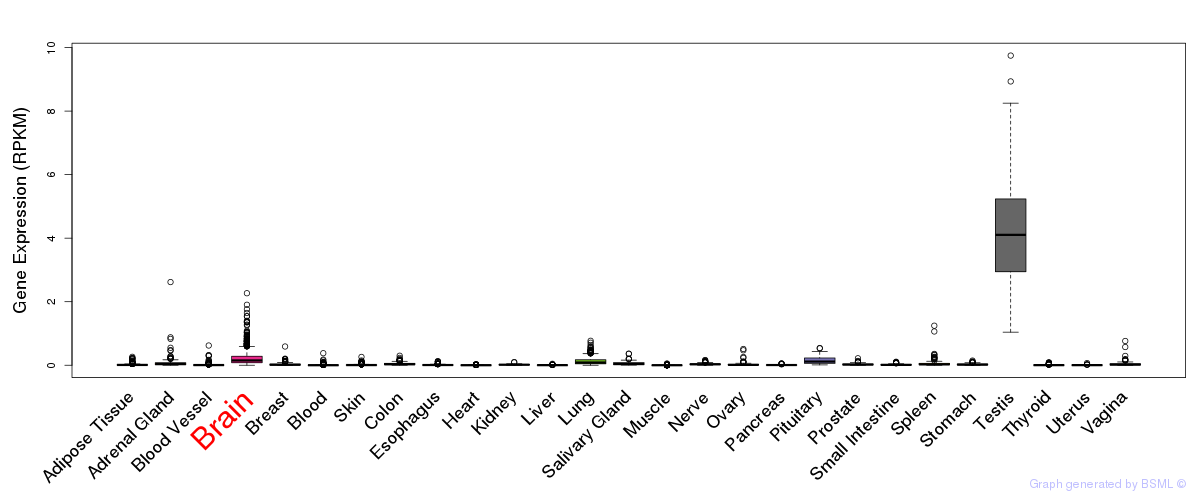
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EFTUD2 | 0.95 | 0.96 |
| VCP | 0.94 | 0.94 |
| FBXO21 | 0.94 | 0.96 |
| DHX8 | 0.94 | 0.95 |
| GANAB | 0.94 | 0.94 |
| EXT2 | 0.94 | 0.95 |
| ZNF207 | 0.93 | 0.93 |
| PRPF4 | 0.93 | 0.94 |
| FBXO38 | 0.93 | 0.93 |
| SND1 | 0.93 | 0.95 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.81 | -0.88 |
| AF347015.31 | -0.79 | -0.86 |
| AF347015.33 | -0.79 | -0.86 |
| FXYD1 | -0.77 | -0.85 |
| AF347015.8 | -0.77 | -0.86 |
| AF347015.21 | -0.77 | -0.89 |
| AF347015.27 | -0.77 | -0.85 |
| MT-CYB | -0.76 | -0.84 |
| IFI27 | -0.75 | -0.81 |
| HIGD1B | -0.74 | -0.82 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004888 | transmembrane receptor activity | TAS | 8861902 | |
| GO:0005515 | protein binding | IPI | 12011067 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007409 | axonogenesis | TAS | neuron, axon, neurite (GO term level: 12) | 8861902 |
| GO:0006917 | induction of apoptosis | TAS | 9796814 | |
| GO:0006915 | apoptosis | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0045786 | negative regulation of cell cycle | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ADORA2B | ADORA2 | adenosine A2b receptor | - | HPRD | 11048721 |
| APPL1 | APPL | DIP13alpha | adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 | - | HPRD,BioGRID | 12011067 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12864730 |
| CASP3 | CPP32 | CPP32B | SCA-1 | caspase 3, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 11248093 |
| CASP9 | APAF-3 | APAF3 | CASPASE-9c | ICE-LAP6 | MCH6 | caspase 9, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 11248093 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | DCC interacts with Fyn. | BIND | 15494732 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | - | HPRD,BioGRID | 11986622 |
| MAZ | PUR1 | Pur-1 | SAF-1 | SAF-2 | ZF87 | ZNF801 | Zif87 | MYC-associated zinc finger protein (purine-binding transcription factor) | - | HPRD,BioGRID | 11527412 |12509857 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | - | HPRD,BioGRID | 12149262 |
| NTN1 | NTN1L | netrin 1 | - | HPRD,BioGRID | 9950216 |12810718 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | Affinity Capture-Western Two-hybrid | BioGRID | 15494733 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | DCC interacts with FAK. | BIND | 15494732 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | FAK interacts with DCC. | BIND | 15494733 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | Affinity Capture-Western | BioGRID | 15494733 |
| SIAH1 | FLJ08065 | HUMSIAH | Siah-1 | Siah-1a | hSIAH1 | seven in absentia homolog 1 (Drosophila) | - | HPRD,BioGRID | 9858595 |
| SIAH1 | FLJ08065 | HUMSIAH | Siah-1 | Siah-1a | hSIAH1 | seven in absentia homolog 1 (Drosophila) | DCC interacts with Siah-1. | BIND | 9334332 |
| SIAH1 | FLJ08065 | HUMSIAH | Siah-1 | Siah-1a | hSIAH1 | seven in absentia homolog 1 (Drosophila) | hSiah1 interacts with DCC cytoplasmic domain. | BIND | 9334332 |
| SIAH2 | hSiah2 | seven in absentia homolog 2 (Drosophila) | - | HPRD,BioGRID | 9858595 |
| SIAH2 | hSiah2 | seven in absentia homolog 2 (Drosophila) | DCC interacts with Siah-2. | BIND | 9334332 |
| SIAH2 | hSiah2 | seven in absentia homolog 2 (Drosophila) | hSiah2 interacts with DCC. | BIND | 9334332 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG COLORECTAL CANCER | 62 | 47 | All SZGR 2.0 genes in this pathway |
| PID NETRIN PATHWAY | 32 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF APOPTOSIS | 58 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | 10 | 5 | All SZGR 2.0 genes in this pathway |
| REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME NETRIN1 SIGNALING | 41 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME DSCAM INTERACTIONS | 11 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A DN | 90 | 61 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER METHYLATED DE NOVO IN CANCER | 88 | 64 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 VS CD2 UP | 66 | 47 | All SZGR 2.0 genes in this pathway |
| HILLION HMGA1 TARGETS | 90 | 71 | All SZGR 2.0 genes in this pathway |
| HILLION HMGA1B TARGETS | 92 | 68 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS HCP WITH H3 UNMETHYLATED | 80 | 50 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES HCP WITH H3K27ME3 | 41 | 30 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-448 | 746 | 753 | 1A,m8 | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-486 | 98 | 104 | m8 | hsa-miR-486 | UCCUGUACUGAGCUGCCCCGAG |
| miR-9 | 487 | 493 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.