Gene Page: DLG1
Summary ?
| GeneID | 1739 |
| Symbol | DLG1 |
| Synonyms | DLGH1|SAP-97|SAP97|dJ1061C18.1.1|hdlg |
| Description | discs large homolog 1, scribble cell polarity complex component |
| Reference | MIM:601014|HGNC:HGNC:2900|Ensembl:ENSG00000075711|HPRD:03007|Vega:OTTHUMG00000047972 |
| Gene type | protein-coding |
| Map location | 3q29 |
| Pascal p-value | 0.017 |
| Fetal beta | -1.399 |
| eGene | Cerebellar Hemisphere Cerebellum Myers' cis & trans |
| Support | PROTEIN CLUSTERING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_PocklingtonH1 G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanNRC CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 1.2622 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DLG1 | chr3 | 196863463 | C | T | NM_001098424 NM_001204386 NM_001204387 NM_001204388 NM_004087 | p.357G>S p.324G>S p.241G>S p.241G>S p.357G>S | missense missense missense missense missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17015843 | chr2 | 79141922 | DLG1 | 1739 | 0.01 | trans | ||
| rs17015845 | chr2 | 79142386 | DLG1 | 1739 | 0 | trans | ||
| rs17015849 | chr2 | 79142711 | DLG1 | 1739 | 0 | trans | ||
| rs17015851 | chr2 | 79143328 | DLG1 | 1739 | 0 | trans | ||
| rs17015854 | chr2 | 79143361 | DLG1 | 1739 | 0 | trans | ||
| rs17015885 | chr2 | 79151081 | DLG1 | 1739 | 0.08 | trans | ||
| rs10219790 | chr13 | 55281771 | DLG1 | 1739 | 0.15 | trans |
Section II. Transcriptome annotation
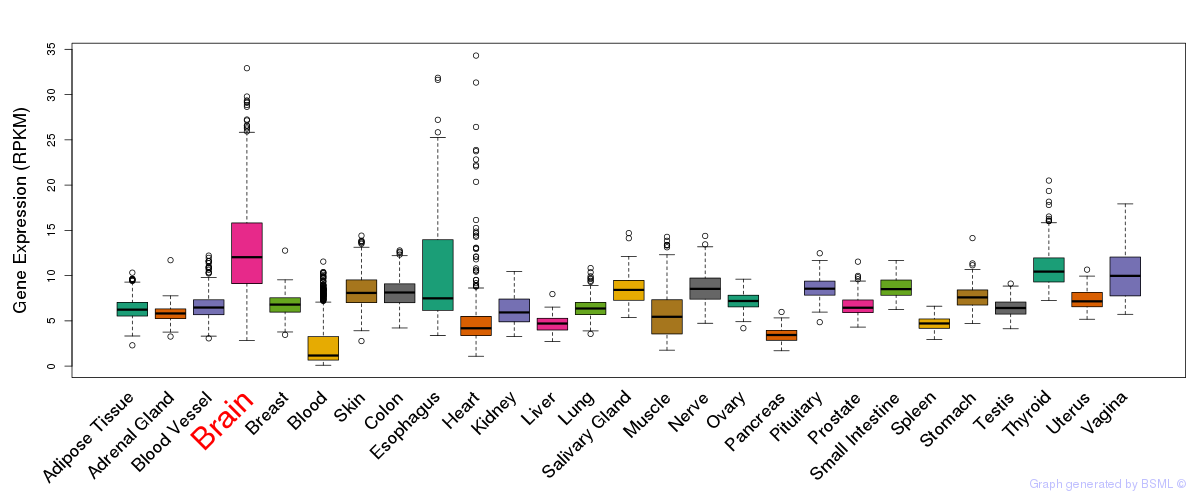
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| GO:0004721 | phosphoprotein phosphatase activity | TAS | 15302935 | |
| GO:0004385 | guanylate kinase activity | TAS | 7937897 | |
| GO:0019902 | phosphatase binding | IPI | 10646847 | |
| GO:0008092 | cytoskeletal protein binding | TAS | 8825652 | |
| GO:0008022 | protein C-terminus binding | IPI | 11274188 | |
| GO:0015459 | potassium channel regulator activity | NAS | 10779557 | |
| GO:0019901 | protein kinase binding | IPI | 9341123 |10779557 |14699157 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001935 | endothelial cell proliferation | IDA | 14699157 | |
| GO:0007163 | establishment or maintenance of cell polarity | TAS | 12766944 | |
| GO:0007015 | actin filament organization | IDA | 14699157 | |
| GO:0016337 | cell-cell adhesion | IDA | 14699157 | |
| GO:0031575 | G1/S transition checkpoint | NAS | 10656683 | |
| GO:0030866 | cortical actin cytoskeleton organization | IDA | 14699157 | |
| GO:0045930 | negative regulation of mitotic cell cycle | IMP | 10656683 | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005789 | endoplasmic reticulum membrane | IEA | - | |
| GO:0005783 | endoplasmic reticulum | IEA | - | |
| GO:0005911 | cell-cell junction | TAS | 7937897 | |
| GO:0005886 | plasma membrane | TAS | 10859302 | |
| GO:0016323 | basolateral plasma membrane | IDA | 8922391 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTN2 | CMD1AA | actinin, alpha 2 | - | HPRD,BioGRID | 12860415 |
| ADAM17 | CD156b | MGC71942 | TACE | cSVP | ADAM metallopeptidase domain 17 | PDZ domain of SAP97 interacts with C-terminal tail of TACE. | BIND | 12668732 |
| ADAM17 | CD156b | MGC71942 | TACE | cSVP | ADAM metallopeptidase domain 17 | - | HPRD,BioGRID | 12668732 |
| AKAP5 | AKAP75 | AKAP79 | H21 | A kinase (PRKA) anchor protein 5 | - | HPRD | 10939335 |
| APBA1 | D9S411E | MINT1 | X11 | X11A | X11ALPHA | amyloid beta (A4) precursor protein-binding, family A, member 1 | Affinity Capture-Western | BioGRID | 14960569 |
| ATP2B2 | PMCA2 | PMCA2a | PMCA2i | ATPase, Ca++ transporting, plasma membrane 2 | PMCA2b interacts with SAP97. | BIND | 11274188 |
| ATP2B2 | PMCA2 | PMCA2a | PMCA2i | ATPase, Ca++ transporting, plasma membrane 2 | - | HPRD,BioGRID | 11274188 |
| ATP2B4 | ATP2B2 | DKFZp686G08106 | DKFZp686M088 | MXRA1 | PMCA4 | PMCA4b | PMCA4x | ATPase, Ca++ transporting, plasma membrane 4 | Reconstituted Complex | BioGRID | 12511555 |
| ATP2B4 | ATP2B2 | DKFZp686G08106 | DKFZp686M088 | MXRA1 | PMCA4 | PMCA4b | PMCA4x | ATPase, Ca++ transporting, plasma membrane 4 | - | HPRD | 11274188 |
| ATP2B4 | ATP2B2 | DKFZp686G08106 | DKFZp686M088 | MXRA1 | PMCA4 | PMCA4b | PMCA4x | ATPase, Ca++ transporting, plasma membrane 4 | PMCA4b interacts with SAP97. This interaction was modeled on a demonstrated interaction between human PMCA4b and rat SAP97. | BIND | 11274188 |
| BEGAIN | KIAA1446 | brain-enriched guanylate kinase-associated homolog (rat) | - | HPRD | 9756850 |
| BTRC | BETA-TRCP | FBW1A | FBXW1 | FBXW1A | FWD1 | MGC4643 | bTrCP | bTrCP1 | betaTrCP | beta-transducin repeat containing | Affinity Capture-Western Reconstituted Complex | BioGRID | 12902344 |
| CACNG2 | MGC138502 | MGC138504 | calcium channel, voltage-dependent, gamma subunit 2 | Affinity Capture-Western | BioGRID | 11140673 |
| CAMK2A | CAMKA | KIAA0968 | calcium/calmodulin-dependent protein kinase II alpha | - | HPRD,BioGRID | 12933808 |
| CASK | CAGH39 | CMG | FLJ22219 | FLJ31914 | LIN2 | MICPCH | TNRC8 | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | Affinity Capture-Western in vitro in vivo Reconstituted Complex Two-hybrid | BioGRID | 10993877 |12151521 |12351654 |14960569 |
| CASK | CAGH39 | CMG | FLJ22219 | FLJ31914 | LIN2 | MICPCH | TNRC8 | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | CASK interacts with DLG1 (SAP97). | BIND | 15729360 |
| CASK | CAGH39 | CMG | FLJ22219 | FLJ31914 | LIN2 | MICPCH | TNRC8 | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | - | HPRD | 10993877 |12351654 |
| CDH1 | Arc-1 | CD324 | CDHE | ECAD | LCAM | UVO | cadherin 1, type 1, E-cadherin (epithelial) | - | HPRD | 9512503 |
| CRIPT | HSPC139 | cysteine-rich PDZ-binding protein | - | HPRD,BioGRID | 12070168 |
| CTNNA1 | CAP102 | FLJ36832 | catenin (cadherin-associated protein), alpha 1, 102kDa | - | HPRD,BioGRID | 9512503 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | - | HPRD | 9512503 |
| DLG2 | DKFZp781D1854 | DKFZp781E0954 | FLJ37266 | MGC131811 | PSD-93 | discs, large homolog 2, chapsyn-110 (Drosophila) | - | HPRD,BioGRID | 12351654 |
| DLG3 | KIAA1232 | MRX | MRX90 | NE-Dlg | NEDLG | SAP102 | discs, large homolog 3 (neuroendocrine-dlg, Drosophila) | - | HPRD,BioGRID | 12351654 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | Affinity Capture-Western | BioGRID | 9286858 |
| DLGAP1 | DAP-1 | DAP-1-ALPHA | DAP-1-BETA | GKAP | MGC88156 | SAPAP1 | hGKAP | discs, large (Drosophila) homolog-associated protein 1 | - | HPRD,BioGRID | 9286858 |
| DLGAP1 | DAP-1 | DAP-1-ALPHA | DAP-1-BETA | GKAP | MGC88156 | SAPAP1 | hGKAP | discs, large (Drosophila) homolog-associated protein 1 | hDLG interacts with DAP-1alpha. | BIND | 9286858 |
| DLGAP1 | DAP-1 | DAP-1-ALPHA | DAP-1-BETA | GKAP | MGC88156 | SAPAP1 | hGKAP | discs, large (Drosophila) homolog-associated protein 1 | DLGAP1 (GKAP) interacts with DLG1 (SAP97). | BIND | 15729360 |
| DLGAP1 | DAP-1 | DAP-1-ALPHA | DAP-1-BETA | GKAP | MGC88156 | SAPAP1 | hGKAP | discs, large (Drosophila) homolog-associated protein 1 | hDLG interacts with DAP-1beta. | BIND | 9286858 |
| DLGAP2 | DAP2 | SAPAP2 | discs, large (Drosophila) homolog-associated protein 2 | hDLG interacts with DAP-2. | BIND | 9286858 |
| DLGAP4 | DAP4 | KIAA0964 | MGC131862 | RP5-977B1.6 | SAPAP4 | discs, large (Drosophila) homolog-associated protein 4 | - | HPRD | 9115257 |
| EPB41 | 4.1R | EL1 | HE | erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) | - | HPRD,BioGRID | 7937897 |
| ERBB4 | HER4 | MGC138404 | p180erbB4 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) | - | HPRD,BioGRID | 12175853 |
| EZR | CVIL | CVL | DKFZp762H157 | FLJ26216 | MGC1584 | VIL2 | ezrin | - | HPRD,BioGRID | 11726633 |
| GDA | CYPIN | GUANASE | KIAA1258 | MGC9982 | NEDASIN | guanine deaminase | - | HPRD | 10595517 |
| GPR124 | DKFZp434C211 | DKFZp434J0911 | FLJ14390 | KIAA1531 | TEM5 | G protein-coupled receptor 124 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 15021905 |
| GPR125 | PGR21 | TEM5L | G protein-coupled receptor 125 | - | HPRD | 15021905 |
| GRIA1 | GLUH1 | GLUR1 | GLURA | HBGR1 | MGC133252 | glutamate receptor, ionotropic, AMPA 1 | - | HPRD,BioGRID | 9677374 |11567040 |
| GRIK2 | EAA4 | GLR6 | GLUK6 | GLUR6 | MGC74427 | MRT6 | glutamate receptor, ionotropic, kainate 2 | - | HPRD,BioGRID | 11279111 |
| GRIN2A | NMDAR2A | NR2A | glutamate receptor, ionotropic, N-methyl D-aspartate 2A | - | HPRD,BioGRID | 12933808 |
| GRIN2B | MGC142178 | MGC142180 | NMDAR2B | NR2B | hNR3 | glutamate receptor, ionotropic, N-methyl D-aspartate 2B | Affinity Capture-Western | BioGRID | 11997254 |
| GUCY1A2 | GC-SA2 | GUC1A2 | guanylate cyclase 1, soluble, alpha 2 | - | HPRD,BioGRID | 11572861 |
| KCNA4 | HBK4 | HK1 | HPCN2 | HUKII | KCNA4L | KCNA8 | KV1.4 | PCN2 | potassium voltage-gated channel, shaker-related subfamily, member 4 | - | HPRD,BioGRID | 12860415 |
| KCNA5 | ATFB7 | HCK1 | HK2 | HPCN1 | KV1.5 | MGC117058 | MGC117059 | PCN1 | potassium voltage-gated channel, shaker-related subfamily, member 5 | Affinity Capture-Western | BioGRID | 12970345 |
| KCNAB1 | AKR6A3 | KCNA1B | KV-BETA-1 | Kvb1.3 | hKvBeta3 | hKvb3 | potassium voltage-gated channel, shaker-related subfamily, beta member 1 | - | HPRD,BioGRID | 9341123 |
| KCNJ10 | BIRK-10 | KCNJ13-PEN | KIR1.2 | KIR4.1 | potassium inwardly-rectifying channel, subfamily J, member 10 | - | HPRD | 9148889 |
| KCNJ12 | FLJ14167 | IRK2 | KCNJN1 | Kir2.2 | Kir2.2v | hIRK | hIRK1 | hkir2.2x | kcnj12x | potassium inwardly-rectifying channel, subfamily J, member 12 | - | HPRD,BioGRID | 5024025 |11181181 |14960569 |
| KCNJ2 | HHBIRK1 | HHIRK1 | IRK1 | KIR2.1 | LQT7 | SQT3 | potassium inwardly-rectifying channel, subfamily J, member 2 | - | HPRD,BioGRID | 11181181 |
| KCNJ4 | HIR | HIRK2 | HRK1 | IRK3 | Kir2.3 | MGC142066 | MGC142068 | potassium inwardly-rectifying channel, subfamily J, member 4 | Affinity Capture-Western Reconstituted Complex | BioGRID | 11181181 |14960569 |15024025 |
| KCNJ6 | BIR1 | GIRK2 | KATP2 | KCNJ7 | KIR3.2 | MGC126596 | hiGIRK2 | potassium inwardly-rectifying channel, subfamily J, member 6 | Reconstituted Complex | BioGRID | 10619846 |
| KIF13B | GAKIN | KIAA0639 | kinesin family member 13B | - | HPRD,BioGRID | 10859302 |
| KIF1B | CMT2 | CMT2A | CMT2A1 | FLJ23699 | HMSNII | KIAA0591 | KIAA1448 | KLP | MGC134844 | kinesin family member 1B | - | HPRD | 12097473 |
| LCK | YT16 | p56lck | pp58lck | lymphocyte-specific protein tyrosine kinase | - | HPRD,BioGRID | 9341123 |
| LIN7A | LIN-7A | LIN7 | MALS-1 | MGC148143 | TIP-33 | VELI1 | lin-7 homolog A (C. elegans) | Affinity Capture-Western | BioGRID | 14960569 |
| LIN7C | FLJ11215 | LIN-7-C | LIN-7C | MALS-3 | MALS3 | VELI3 | lin-7 homolog C (C. elegans) | Affinity Capture-Western | BioGRID | 14960569 |
| LRP2 | DBS | gp330 | low density lipoprotein-related protein 2 | - | HPRD | 12713445 |
| LRRC1 | FLJ10775 | FLJ11834 | LANO | dJ523E19.1 | leucine rich repeat containing 1 | Two-hybrid | BioGRID | 11440998 |
| MAP1A | FLJ77111 | MAP1L | MTAP1A | microtubule-associated protein 1A | - | HPRD | 9786987 |
| MRPS34 | MGC2616 | MRP-S12 | MRP-S34 | MRPS12 | mitochondrial ribosomal protein S34 | DLG1 (hDLG) interacts with MRPS34 (MRP-S34). | BIND | 12507520 |
| MYO6 | DFNA22 | DFNB37 | KIAA0389 | myosin VI | - | HPRD | 12050163 |
| PBK | FLJ14385 | Nori-3 | SPK | TOPK | PDZ binding kinase | PBK interacts with DLG1 (SAP97). This interaction was modelled on a demonstrated interaction between PBK from an unspecified species and human DLG1 (SAP97). | BIND | 15729360 |
| PBK | FLJ14385 | Nori-3 | SPK | TOPK | PDZ binding kinase | - | HPRD | 10779557 |
| SEMA4C | FLJ20369 | KIAA1739 | M-SEMA-F | MGC126382 | MGC126383 | SEMACL1 | SEMAF | SEMAI | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C | - | HPRD | 11134026 |
| TANC1 | KIAA1728 | ROLSB | TANC | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 | - | HPRD | 15673434 |
| TJAP1 | DKFZp686F06131 | PILT | TJP4 | tight junction associated protein 1 (peripheral) | - | HPRD,BioGRID | 11602598 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| PID CDC42 PATHWAY | 70 | 51 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN NASCENT AJ PATHWAY | 39 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAFFICKING OF AMPA RECEPTORS | 28 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | 31 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | 11 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME L1CAM INTERACTIONS | 86 | 62 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| LIU CMYB TARGETS UP | 165 | 106 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS UP | 112 | 68 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| CHOW RASSF1 TARGETS UP | 27 | 17 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 XPCS DN | 88 | 71 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 8HR UP | 105 | 73 | All SZGR 2.0 genes in this pathway |
| APRELIKOVA BRCA1 TARGETS | 49 | 33 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX UP | 89 | 59 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 12HR UP | 111 | 68 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA AND UV RADIATION | 88 | 65 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| BOHN PRIMARY IMMUNODEFICIENCY SYNDROM UP | 47 | 30 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |