Gene Page: DRD4
Summary ?
| GeneID | 1815 |
| Symbol | DRD4 |
| Synonyms | D4DR |
| Description | dopamine receptor D4 |
| Reference | MIM:126452|HGNC:HGNC:3025|Ensembl:ENSG00000069696|HPRD:00541|Vega:OTTHUMG00000133312 |
| Gene type | protein-coding |
| Map location | 11p15.5 |
| Pascal p-value | 0.357 |
| Sherlock p-value | 0.764 |
| Fetal beta | 0.027 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| ADT:Sun_2012 | Systematic Investigation of Antipsychotic Drugs and Their Targets | A total of 382 drug-target associations involving 43 antipsychotic drugs and 49 target genes. | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 3 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizotypy,schizophrenias,schizotypal | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 9 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 31 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16903386 | chr5 | 88159284 | DRD4 | 1815 | 0.11 | trans | ||
| rs7503446 | chr17 | 37246066 | DRD4 | 1815 | 0.01 | trans |
Section II. Transcriptome annotation
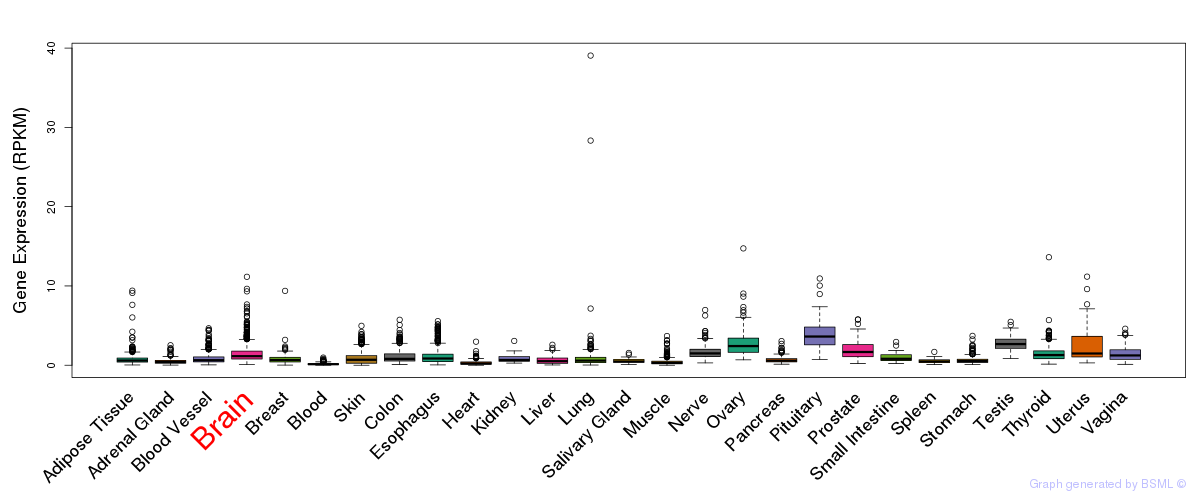
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TBR1 | 0.92 | 0.79 |
| FAM59A | 0.92 | 0.90 |
| SLC35F2 | 0.91 | 0.81 |
| PBX1 | 0.90 | 0.88 |
| OSBPL10 | 0.90 | 0.85 |
| STT3B | 0.89 | 0.90 |
| GPD1L | 0.88 | 0.89 |
| DAP | 0.88 | 0.69 |
| TOX | 0.88 | 0.81 |
| LIMCH1 | 0.88 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SERPINB6 | -0.56 | -0.66 |
| C5orf53 | -0.53 | -0.69 |
| TNFSF12 | -0.53 | -0.58 |
| HSD17B14 | -0.53 | -0.73 |
| IFI27 | -0.52 | -0.80 |
| FXYD1 | -0.52 | -0.82 |
| AC018755.7 | -0.51 | -0.65 |
| AC007405.8 | -0.51 | -0.60 |
| PTH1R | -0.51 | -0.68 |
| AIFM3 | -0.51 | -0.70 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004952 | dopamine receptor activity | IEA | Neurotransmitter, dopamine (GO term level: 8) | - |
| GO:0004952 | dopamine receptor activity | NAS | Neurotransmitter, dopamine (GO term level: 8) | 8746407 |
| GO:0001593 | dopamine D4 receptor activity | IEA | dopamine (GO term level: 10) | - |
| GO:0001591 | dopamine receptor coupled via Gi/Go | TAS | Neurotransmitter, dopamine (GO term level: 9) | 9457173 |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0008144 | drug binding | IDA | 1840645 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0060080 | regulation of inhibitory postsynaptic membrane potential | IEA | neuron, Synap (GO term level: 10) | - |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 8413587 |
| GO:0007195 | dopamine receptor, adenylate cyclase inhibiting pathway | IEA | dopamine (GO term level: 12) | - |
| GO:0007195 | dopamine receptor, adenylate cyclase inhibiting pathway | TAS | dopamine (GO term level: 12) | 9457173 |
| GO:0042053 | regulation of dopamine metabolic process | IEA | dopamine (GO term level: 9) | - |
| GO:0001975 | response to amphetamine | IEA | - | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IEA | - | |
| GO:0007194 | negative regulation of adenylate cyclase activity | TAS | 9457173 | |
| GO:0008344 | adult locomotory behavior | IEA | - | |
| GO:0006874 | cellular calcium ion homeostasis | IC | 7921596 | |
| GO:0014075 | response to amine stimulus | IDA | 16839358 | |
| GO:0042596 | fear response | IEA | - | |
| GO:0048148 | behavioral response to cocaine | IEA | - | |
| GO:0050709 | negative regulation of protein secretion | IDA | 16839358 | |
| GO:0051927 | negative regulation of calcium ion transport via voltage-gated calcium channel | IDA | 7921596 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | NAS | 8746407 | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | TAS | 1319557 | |
| GO:0005887 | integral to plasma membrane | TAS | 1319557 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX UP | 83 | 66 | All SZGR 2.0 genes in this pathway |
| HOQUE METHYLATED IN CANCER | 56 | 45 | All SZGR 2.0 genes in this pathway |
| MUELLER COMMON TARGETS OF AML FUSIONS DN | 31 | 21 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 4 | 15 | 11 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 DN | 157 | 106 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |