Gene Page: SLC26A2
Summary ?
| GeneID | 1836 |
| Symbol | SLC26A2 |
| Synonyms | D5S1708|DTD|DTDST|EDM4|MST153|MSTP157 |
| Description | solute carrier family 26 member 2 |
| Reference | MIM:606718|HGNC:HGNC:10994|Ensembl:ENSG00000155850|HPRD:05990|Vega:OTTHUMG00000130054 |
| Gene type | protein-coding |
| Map location | 5q32 |
| Pascal p-value | 0.244 |
| Fetal beta | 0.902 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00459 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01718 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20540378 | 5 | 149340137 | SLC26A2 | 1.5E-8 | -0.007 | 5.7E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6874540 | chr5 | 149911669 | SLC26A2 | 1836 | 0.13 | cis | ||
| rs2273232 | chr5 | 149912526 | SLC26A2 | 1836 | 0.13 | cis | ||
| rs17131472 | chr1 | 91962966 | SLC26A2 | 1836 | 0.01 | trans | ||
| rs1473237 | chr1 | 191045246 | SLC26A2 | 1836 | 0.11 | trans | ||
| rs2115055 | chr2 | 119478688 | SLC26A2 | 1836 | 0.09 | trans | ||
| rs2922180 | chr3 | 125280472 | SLC26A2 | 1836 | 7.742E-5 | trans | ||
| rs2976809 | chr3 | 125451738 | SLC26A2 | 1836 | 3.095E-4 | trans | ||
| rs16889842 | chr4 | 9872012 | SLC26A2 | 1836 | 0.02 | trans | ||
| snp_a-1976228 | 0 | SLC26A2 | 1836 | 0.08 | trans | |||
| rs2732167 | chr4 | 88706292 | SLC26A2 | 1836 | 0.03 | trans | ||
| rs313946 | chr4 | 113975904 | SLC26A2 | 1836 | 0 | trans | ||
| rs10050805 | chr5 | 30805352 | SLC26A2 | 1836 | 0.05 | trans | ||
| rs17056435 | chr5 | 158453782 | SLC26A2 | 1836 | 0 | trans | ||
| rs17135501 | chr6 | 2704474 | SLC26A2 | 1836 | 0.01 | trans | ||
| rs17135521 | chr6 | 2707631 | SLC26A2 | 1836 | 0.01 | trans | ||
| snp_a-1787926 | 0 | SLC26A2 | 1836 | 0.01 | trans | |||
| rs6455226 | chr6 | 67930295 | SLC26A2 | 1836 | 0.05 | trans | ||
| rs8192621 | chr6 | 132966206 | SLC26A2 | 1836 | 0.03 | trans | ||
| rs17065063 | chr6 | 136207301 | SLC26A2 | 1836 | 0.17 | trans | ||
| rs2398707 | chr7 | 2113428 | SLC26A2 | 1836 | 0.16 | trans | ||
| rs3778951 | chr7 | 2125319 | SLC26A2 | 1836 | 0.16 | trans | ||
| rs3778957 | chr7 | 2127710 | SLC26A2 | 1836 | 0.16 | trans | ||
| rs17156987 | chr7 | 28704217 | SLC26A2 | 1836 | 0.2 | trans | ||
| rs2286089 | chr7 | 38789903 | SLC26A2 | 1836 | 0.09 | trans | ||
| rs2430483 | chr7 | 104548631 | SLC26A2 | 1836 | 0.03 | trans | ||
| rs888577 | chr8 | 239323 | SLC26A2 | 1836 | 0.17 | trans | ||
| rs2515434 | chr8 | 6374892 | SLC26A2 | 1836 | 0.05 | trans | ||
| rs2442623 | chr8 | 6376337 | SLC26A2 | 1836 | 0.05 | trans | ||
| rs2515437 | chr8 | 6377140 | SLC26A2 | 1836 | 0.05 | trans | ||
| rs1960369 | chr8 | 6379235 | SLC26A2 | 1836 | 0.01 | trans | ||
| rs2515463 | chr8 | 6384137 | SLC26A2 | 1836 | 0.12 | trans | ||
| rs2919377 | chr8 | 32573740 | SLC26A2 | 1836 | 0.11 | trans | ||
| rs2253777 | chr9 | 93467893 | SLC26A2 | 1836 | 0.12 | trans | ||
| rs6478086 | chr9 | 117273020 | SLC26A2 | 1836 | 0.03 | trans | ||
| rs998410 | chr9 | 117622673 | SLC26A2 | 1836 | 1.521E-5 | trans | ||
| rs9423711 | chr10 | 2574802 | SLC26A2 | 1836 | 0.17 | trans | ||
| rs4514358 | chr10 | 43861923 | SLC26A2 | 1836 | 0.04 | trans | ||
| rs2116478 | chr11 | 83362424 | SLC26A2 | 1836 | 0.05 | trans | ||
| rs2098090 | chr12 | 4770747 | SLC26A2 | 1836 | 0.09 | trans | ||
| rs10747972 | 0 | SLC26A2 | 1836 | 0.02 | trans | |||
| rs995817 | chr12 | 67557935 | SLC26A2 | 1836 | 0 | trans | ||
| rs11112163 | chr12 | 105072508 | SLC26A2 | 1836 | 0.02 | trans | ||
| rs9551590 | chr13 | 29589464 | SLC26A2 | 1836 | 0.01 | trans | ||
| rs9529974 | chr13 | 72821935 | SLC26A2 | 1836 | 0.05 | trans | ||
| rs11840833 | chr13 | 94378306 | SLC26A2 | 1836 | 0.01 | trans | ||
| rs17381373 | chr13 | 109002943 | SLC26A2 | 1836 | 0.18 | trans | ||
| rs10142086 | chr14 | 35135892 | SLC26A2 | 1836 | 0.08 | trans | ||
| rs1956517 | chr14 | 37738426 | SLC26A2 | 1836 | 0.1 | trans | ||
| rs10498433 | chr14 | 52289530 | SLC26A2 | 1836 | 0 | trans | ||
| rs2093759 | chr14 | 97171024 | SLC26A2 | 1836 | 0.09 | trans | ||
| rs17244419 | chr14 | 97171074 | SLC26A2 | 1836 | 0 | trans | ||
| rs16976022 | chr15 | 55454260 | SLC26A2 | 1836 | 0.2 | trans | ||
| rs16976029 | chr15 | 55455268 | SLC26A2 | 1836 | 0.13 | trans | ||
| rs16977927 | chr17 | 72154232 | SLC26A2 | 1836 | 0.06 | trans | ||
| rs16940550 | chr18 | 13442256 | SLC26A2 | 1836 | 0.07 | trans | ||
| rs11873703 | chr18 | 61448702 | SLC26A2 | 1836 | 0 | trans | ||
| rs9951107 | chr18 | 74437994 | SLC26A2 | 1836 | 0.07 | trans | ||
| rs1075540 | chr22 | 25254707 | SLC26A2 | 1836 | 0.09 | trans | ||
| rs6000401 | chr22 | 37149335 | SLC26A2 | 1836 | 0.14 | trans | ||
| rs5991662 | chrX | 43335796 | SLC26A2 | 1836 | 0.06 | trans |
Section II. Transcriptome annotation
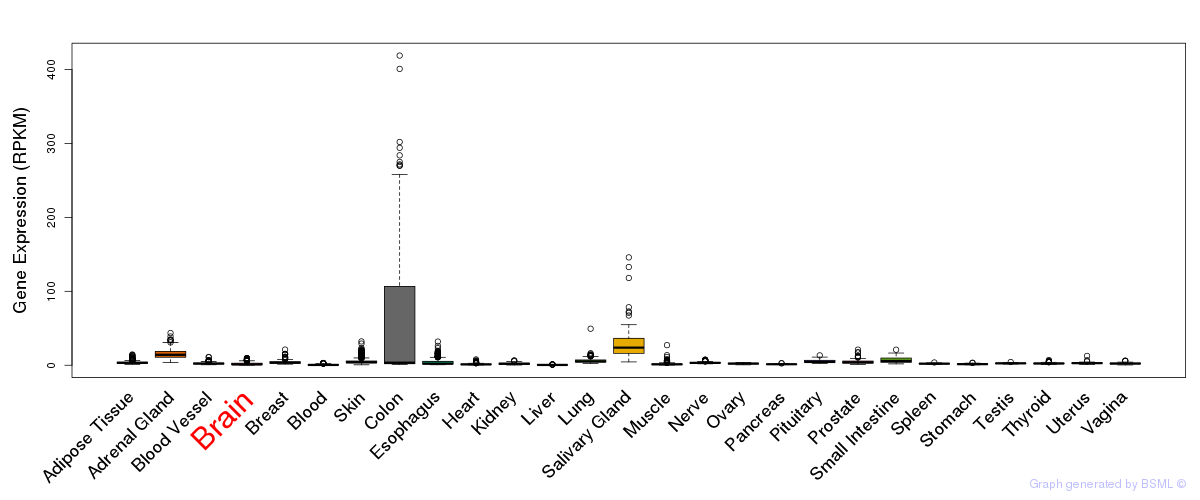
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC024909.1 | 0.98 | 0.98 |
| CACNB2 | 0.85 | 0.86 |
| CNNM1 | 0.84 | 0.83 |
| PRRT3 | 0.83 | 0.85 |
| SLC35F3 | 0.83 | 0.84 |
| KCNS2 | 0.83 | 0.83 |
| CX3CL1 | 0.82 | 0.84 |
| GPR61 | 0.82 | 0.85 |
| EGR2 | 0.82 | 0.79 |
| NALCN | 0.81 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RBMX2 | -0.43 | -0.51 |
| SNHG12 | -0.42 | -0.52 |
| GTF3C6 | -0.42 | -0.42 |
| EXOSC8 | -0.41 | -0.40 |
| RPL38 | -0.41 | -0.56 |
| RPL37 | -0.41 | -0.58 |
| RPS20 | -0.41 | -0.52 |
| RPL23A | -0.41 | -0.49 |
| RPS13P2 | -0.40 | -0.43 |
| RPS27A | -0.40 | -0.49 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005215 | transporter activity | IEA | - | |
| GO:0008271 | secondary active sulfate transmembrane transporter activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008272 | sulfate transport | TAS | 7923357 | |
| GO:0006810 | transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005624 | membrane fraction | TAS | 8528239 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 8528239 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME SLC MEDIATED TRANSMEMBRANE TRANSPORT | 241 | 157 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | 94 | 65 | All SZGR 2.0 genes in this pathway |
| WATANABE COLON CANCER MSI VS MSS DN | 81 | 42 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS UP | 269 | 146 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL DN | 101 | 66 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| WILLIAMS ESR1 TARGETS UP | 26 | 16 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 480 MCF10A | 39 | 20 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 2 | 114 | 76 | All SZGR 2.0 genes in this pathway |
| CHEOK RESPONSE TO MERCAPTOPURINE UP | 13 | 10 | All SZGR 2.0 genes in this pathway |
| NELSON RESPONSE TO ANDROGEN UP | 86 | 61 | All SZGR 2.0 genes in this pathway |
| TRAYNOR RETT SYNDROM DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| NIELSEN SCHWANNOMA UP | 18 | 16 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| MASSARWEH RESPONSE TO ESTRADIOL | 61 | 47 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN | 202 | 132 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K9ME3 DN | 120 | 71 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY DN | 58 | 39 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G23 UP | 52 | 35 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| KYNG RESPONSE TO H2O2 VIA ERCC6 UP | 40 | 30 | All SZGR 2.0 genes in this pathway |
| STAMBOLSKY RESPONSE TO VITAMIN D3 UP | 84 | 48 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |