Gene Page: DTNA
Summary ?
| GeneID | 1837 |
| Symbol | DTNA |
| Synonyms | D18S892E|DRP3|DTN|DTN-A|LVNC1 |
| Description | dystrobrevin alpha |
| Reference | MIM:601239|HGNC:HGNC:3057|Ensembl:ENSG00000134769|HPRD:03141|Vega:OTTHUMG00000132309 |
| Gene type | protein-coding |
| Map location | 18q12 |
| Pascal p-value | 0.033 |
| Sherlock p-value | 0.951 |
| Fetal beta | -1.909 |
| eGene | Cerebellar Hemisphere Meta |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.4164 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
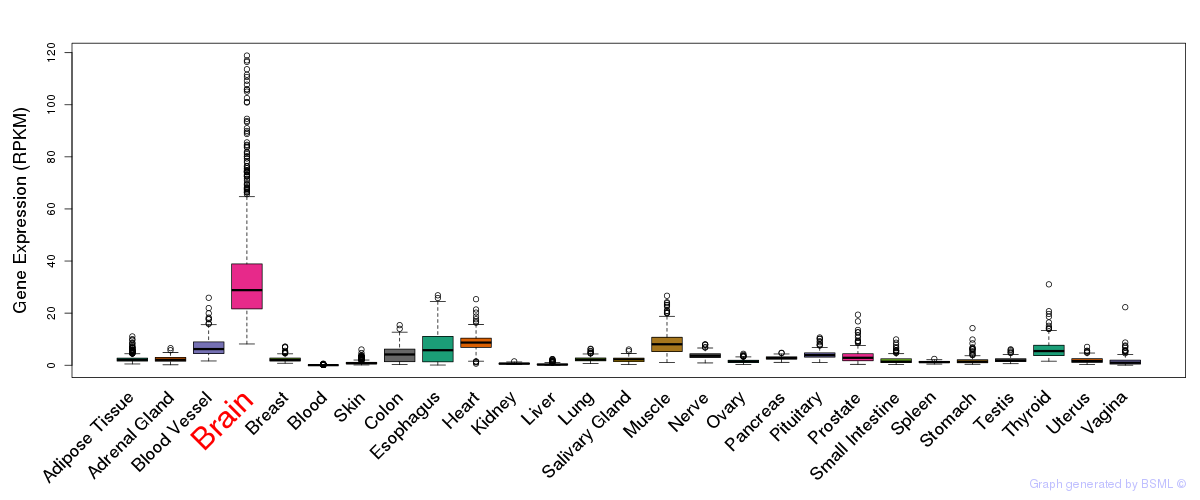
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HRH2 | 0.87 | 0.89 |
| PITPNM2 | 0.87 | 0.87 |
| RHOBTB2 | 0.87 | 0.86 |
| PIP5K1C | 0.86 | 0.86 |
| OSBP2 | 0.86 | 0.88 |
| MGAT5B | 0.86 | 0.87 |
| ABR | 0.86 | 0.87 |
| BAI2 | 0.86 | 0.87 |
| ZDHHC8 | 0.86 | 0.85 |
| BAIAP2 | 0.85 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DBI | -0.51 | -0.57 |
| AF347015.21 | -0.50 | -0.43 |
| PECI | -0.50 | -0.54 |
| NSBP1 | -0.50 | -0.54 |
| GNG11 | -0.50 | -0.52 |
| RAB13 | -0.49 | -0.57 |
| AP002478.3 | -0.48 | -0.49 |
| MYL12A | -0.48 | -0.54 |
| AL139819.3 | -0.47 | -0.49 |
| C1orf54 | -0.47 | -0.49 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 11353857 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007274 | neuromuscular synaptic transmission | TAS | neuron, Synap (GO term level: 7) | 9119373 |
| GO:0007165 | signal transduction | TAS | 10767327 | |
| GO:0006941 | striated muscle contraction | TAS | 10767327 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| DMD | BMD | CMD3B | DXS142 | DXS164 | DXS206 | DXS230 | DXS239 | DXS268 | DXS269 | DXS270 | DXS272 | dystrophin | - | HPRD,BioGRID | 9356463 |
| DRP2 | MGC133255 | dystrophin related protein 2 | - | HPRD,BioGRID | 10767429 |
| DTNBP1 | DBND | DKFZp564K192 | FLJ30031 | HPS7 | MGC20210 | My031 | SDY | dystrobrevin binding protein 1 | - | HPRD | 11316798 |
| KCNJ12 | FLJ14167 | IRK2 | KCNJN1 | Kir2.2 | Kir2.2v | hIRK | hIRK1 | hkir2.2x | kcnj12x | potassium inwardly-rectifying channel, subfamily J, member 12 | - | HPRD,BioGRID | 15024025 |
| SNTA1 | SNT1 | TACIP1 | dJ1187J4.5 | syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component) | - | HPRD | 12206805 |
| SNTA1 | SNT1 | TACIP1 | dJ1187J4.5 | syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component) | - | HPRD | 11069112 |12206805|12206805 |
| SNTB1 | 59-DAP | A1B | BSYN2 | DAPA1B | FLJ22442 | MGC111389 | SNT2 | SNT2B1 | TIP-43 | syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) | - | HPRD | 10545507 |
| SNTB1 | 59-DAP | A1B | BSYN2 | DAPA1B | FLJ22442 | MGC111389 | SNT2 | SNT2B1 | TIP-43 | syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) | - | HPRD | 7844150 |10545507 |11069112|10545507 |
| SNTB2 | D16S2531E | EST25263 | SNT2B2 | SNT3 | SNTL | syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) | - | HPRD | 12206805 |
| SNTG1 | G1SYN | SYN4 | syntrophin, gamma 1 | - | HPRD | 10747910 |
| SNTG2 | G2SYN | MGC133174 | SYN5 | syntrophin, gamma 2 | - | HPRD | 10747910 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD | 9701558 |
| SYNC | MGC149625 | MGC149626 | SYNC1 | SYNCOILIN | syncoilin, intermediate filament protein | - | HPRD | 11053421 |
| SYNM | DMN | KIAA0353 | SYN | synemin, intermediate filament protein | - | HPRD,BioGRID | 11353857 |
| UTRN | DMDL | DRP | DRP1 | FLJ23678 | utrophin | - | HPRD,BioGRID | 9701558 |12034776 |
| UTRN | DMDL | DRP | DRP1 | FLJ23678 | utrophin | - | HPRD | 9701558 |10767429 |12034776 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE DN | 84 | 53 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC2 TARGETS DN | 133 | 77 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL DN | 118 | 79 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP | 171 | 112 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA BY DMOG UP | 130 | 85 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A UP | 191 | 128 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH OCT4 TARGETS | 290 | 172 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH NOS TARGETS | 179 | 105 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE UP | 86 | 57 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA MS UP | 48 | 32 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX UP | 83 | 66 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| SAFFORD T LYMPHOCYTE ANERGY | 87 | 54 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN | 202 | 132 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 DN | 228 | 114 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |
| PHESSE TARGETS OF APC AND MBD2 DN | 13 | 11 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS UP | 116 | 87 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 1146 | 1152 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 1146 | 1152 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-128 | 887 | 893 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-135 | 772 | 778 | 1A | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA | ||||
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-140 | 931 | 938 | 1A,m8 | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-143 | 2149 | 2155 | m8 | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
| miR-146 | 2086 | 2093 | 1A,m8 | hsa-miR-146a | UGAGAACUGAAUUCCAUGGGUU |
| hsa-miR-146bbrain | UGAGAACUGAAUUCCAUAGGCU | ||||
| miR-148/152 | 3149 | 3155 | 1A | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| hsa-miR-148a | UCAGUGCACUACAGAACUUUGU | ||||
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-183 | 940 | 947 | 1A,m8 | hsa-miR-183 | UAUGGCACUGGUAGAAUUCACUG |
| miR-185 | 2675 | 2681 | 1A | hsa-miR-185brain | UGGAGAGAAAGGCAGUUC |
| miR-19 | 2100 | 2107 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA | ||||
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA | ||||
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-22 | 213 | 219 | 1A | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-24 | 3122 | 3128 | 1A | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-27 | 887 | 894 | 1A,m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC | ||||
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-299-5p | 930 | 936 | 1A | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
| miR-30-5p | 439 | 445 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-320 | 1078 | 1085 | 1A,m8 | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-365 | 3317 | 3323 | m8 | hsa-miR-365 | UAAUGCCCCUAAAAAUCCUUAU |
| miR-376c | 3101 | 3107 | 1A | hsa-miR-376c | AACAUAGAGGAAAUUCCACG |
| miR-378 | 2917 | 2923 | 1A | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
| hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU | ||||
| miR-410 | 3577 | 3583 | m8 | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-448 | 3115 | 3121 | m8 | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-9 | 2065 | 2071 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.