Gene Page: EN1
Summary ?
| GeneID | 2019 |
| Symbol | EN1 |
| Synonyms | - |
| Description | engrailed homeobox 1 |
| Reference | MIM:131290|HGNC:HGNC:3342|Ensembl:ENSG00000163064|HPRD:08835|Vega:OTTHUMG00000131401 |
| Gene type | protein-coding |
| Map location | 2q14.2 |
| Pascal p-value | 0.015 |
| Fetal beta | -0.049 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0004 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00755 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg05270938 | 2 | 119616568 | EN1 | 6.07E-8 | -0.018 | 1.53E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10994209 | chr10 | 61877705 | EN1 | 2019 | 0.17 | trans |
Section II. Transcriptome annotation
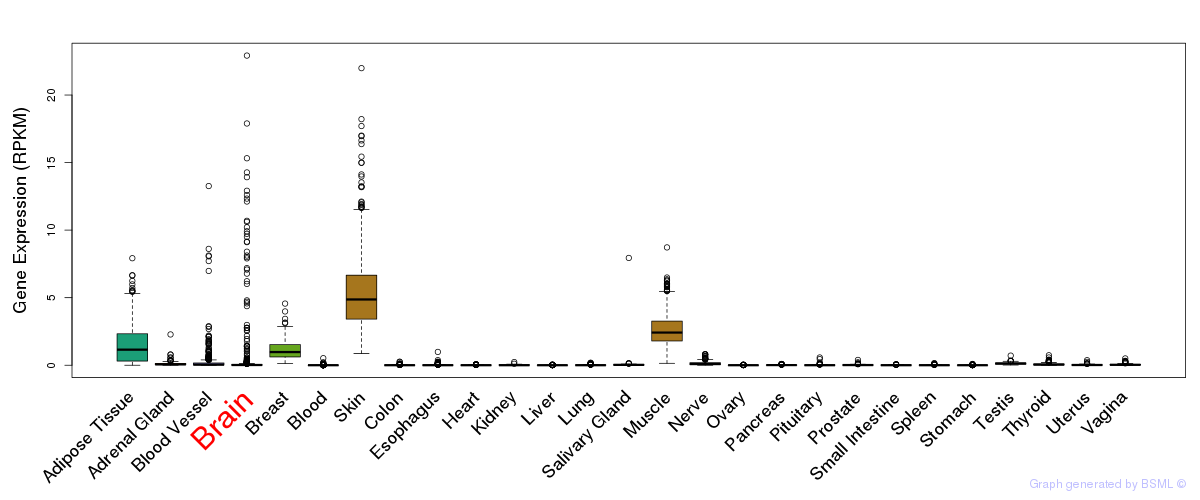
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ABCB1 | 0.87 | 0.88 |
| TPCN1 | 0.86 | 0.89 |
| KANK1 | 0.86 | 0.87 |
| ATP1A2 | 0.86 | 0.87 |
| SFXN5 | 0.86 | 0.84 |
| GPR37L1 | 0.85 | 0.83 |
| SEMA3G | 0.85 | 0.85 |
| NACC2 | 0.85 | 0.91 |
| SLC6A12 | 0.85 | 0.90 |
| PRELP | 0.84 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| POLB | -0.67 | -0.77 |
| MED19 | -0.67 | -0.72 |
| FRG1 | -0.66 | -0.79 |
| TRNAU1AP | -0.66 | -0.73 |
| OCIAD2 | -0.66 | -0.73 |
| ING4 | -0.65 | -0.70 |
| DUSP12 | -0.65 | -0.68 |
| ALKBH2 | -0.65 | -0.75 |
| COMMD3 | -0.65 | -0.76 |
| NR2C2AP | -0.64 | -0.72 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | IEA | - | |
| GO:0005249 | voltage-gated potassium channel activity | IEA | - | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0048666 | neuron development | IEA | neuron (GO term level: 9) | - |
| GO:0030917 | midbrain-hindbrain boundary development | IEA | Brain (GO term level: 10) | - |
| GO:0030902 | hindbrain development | IEA | Brain (GO term level: 8) | - |
| GO:0030901 | midbrain development | IEA | Brain (GO term level: 8) | - |
| GO:0001501 | skeletal system development | TAS | 8684466 | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0009954 | proximal/distal pattern formation | IEA | - | |
| GO:0009953 | dorsal/ventral pattern formation | IEA | - | |
| GO:0009653 | anatomical structure morphogenesis | TAS | 8094370 | |
| GO:0006813 | potassium ion transport | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0043473 | pigmentation | IEA | - | |
| GO:0035115 | embryonic forelimb morphogenesis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0008076 | voltage-gated potassium channel complex | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DOANE BREAST CANCER ESR1 DN | 48 | 28 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A UP | 191 | 128 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES CORE NINE CORRELATED | 100 | 68 | All SZGR 2.0 genes in this pathway |
| LEIN MIDBRAIN MARKERS | 82 | 55 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BRAIN UP | 39 | 20 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| BOUDOUKHA BOUND BY IGF2BP2 | 111 | 59 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP | 297 | 194 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-128 | 754 | 760 | m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-181 | 478 | 484 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-29 | 200 | 206 | 1A | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-338 | 350 | 356 | 1A | hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA |
| miR-369-3p | 394 | 400 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU | ||||
| miR-374 | 403 | 410 | 1A,m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG | ||||
| miR-491 | 671 | 677 | 1A | hsa-miR-491brain | AGUGGGGAACCCUUCCAUGAGGA |
| miR-9 | 706 | 712 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.