Gene Page: GABBR1
Summary ?
| GeneID | 2550 |
| Symbol | GABBR1 |
| Synonyms | GABABR1|GABBR1-3|GB1|GPRC3A |
| Description | gamma-aminobutyric acid type B receptor subunit 1 |
| Reference | MIM:603540|HGNC:HGNC:4070|Ensembl:ENSG00000204681|HPRD:04644|Vega:OTTHUMG00000031095 |
| Gene type | protein-coding |
| Map location | 6p21.31 |
| Pascal p-value | 1E-12 |
| Sherlock p-value | 0.351 |
| Fetal beta | -0.847 |
| DMG | 1 (# studies) |
| Support | LIGAND GATED ION SIGNALING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0246 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg21552290 | 6 | 29578423 | GABBR1 | 8.92E-5 | 0.444 | 0.027 | DMG:Wockner_2014 |
| cg24628013 | 6 | 29580367 | GABBR1 | 2.66E-4 | 0.353 | 0.038 | DMG:Wockner_2014 |
| cg17806418 | 6 | 29599319 | GABBR1 | 5.478E-4 | 0.398 | 0.049 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
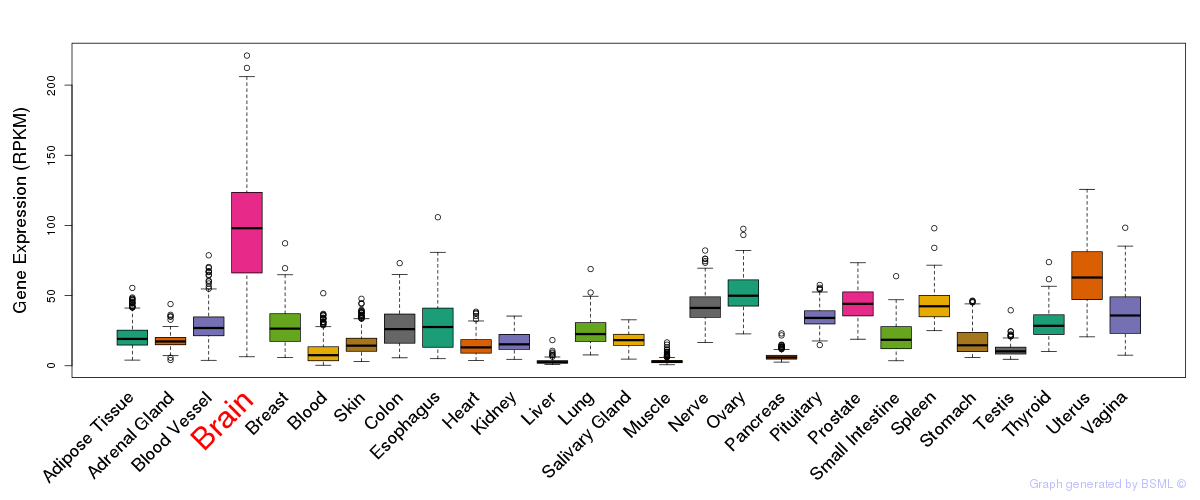
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PREPL | 0.95 | 0.93 |
| NSF | 0.94 | 0.94 |
| ATP6V1B2 | 0.93 | 0.93 |
| SYNJ1 | 0.92 | 0.92 |
| GLS | 0.92 | 0.92 |
| NCOA7 | 0.91 | 0.90 |
| GABRA1 | 0.91 | 0.88 |
| SLC3A1 | 0.90 | 0.92 |
| ATP6V0A1 | 0.90 | 0.94 |
| EFR3A | 0.90 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.44 | -0.51 |
| EFEMP2 | -0.42 | -0.43 |
| RAB13 | -0.41 | -0.50 |
| NME4 | -0.41 | -0.53 |
| RAB34 | -0.40 | -0.45 |
| IRF7 | -0.40 | -0.51 |
| CLEC3B | -0.39 | -0.50 |
| WDR86 | -0.38 | -0.34 |
| AC006276.2 | -0.37 | -0.34 |
| C1orf61 | -0.37 | -0.44 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004965 | GABA-B receptor activity | IEA | GABA, glutamate (GO term level: 7) | - |
| GO:0004965 | GABA-B receptor activity | TAS | GABA, glutamate (GO term level: 7) | 9069281 |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0004930 | G-protein coupled receptor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007214 | gamma-aminobutyric acid signaling pathway | TAS | GABA, Neurotransmitter (GO term level: 8) | 9069281 |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IEA | - | |
| GO:0006464 | protein modification process | IEA | - | |
| GO:0007194 | negative regulation of adenylate cyclase activity | TAS | 9069281 | |
| GO:0007165 | signal transduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 9753614 | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | 15 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | 25 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA B RECEPTOR ACTIVATION | 38 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA RECEPTOR ACTIVATION | 52 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME POTASSIUM CHANNELS | 98 | 68 | All SZGR 2.0 genes in this pathway |
| REACTOME INWARDLY RECTIFYING K CHANNELS | 31 | 20 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA PROGENITOR UP | 39 | 25 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH REARRANGEMENTS UP | 48 | 34 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| MOREAUX B LYMPHOCYTE MATURATION BY TACI UP | 92 | 58 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM UP | 84 | 58 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE NOT BY 4NQO IN WS | 40 | 26 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE NOT BY GAMMA IN OLD | 31 | 19 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE NOT BY GAMMA IN WS | 33 | 22 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE DN | 21 | 12 | All SZGR 2.0 genes in this pathway |
| YOKOE CANCER TESTIS ANTIGENS | 38 | 22 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN | 209 | 137 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS FIBROBLAST UP | 84 | 60 | All SZGR 2.0 genes in this pathway |
| GUTIERREZ WALDENSTROEMS MACROGLOBULINEMIA 1 UP | 9 | 5 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS DURATION CORR DN | 146 | 90 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM SENSITIVE GENES | 90 | 54 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| LE NEURONAL DIFFERENTIATION UP | 18 | 13 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL DN | 146 | 88 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL DN | 214 | 124 | All SZGR 2.0 genes in this pathway |