Gene Page: LSM3
Summary ?
| GeneID | 27258 |
| Symbol | LSM3 |
| Synonyms | SMX4|USS2|YLR438C |
| Description | LSM3 homolog, U6 small nuclear RNA and mRNA degradation associated |
| Reference | MIM:607283|HGNC:HGNC:17874|HPRD:06283| |
| Gene type | protein-coding |
| Map location | 3p25.1 |
| Pascal p-value | 0.843 |
| Sherlock p-value | 0.661 |
| Fetal beta | 0.153 |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.006 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.3063 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| LSM3 | chr3 | 14239557 | A | G | NM_014463 | p.84M>V | missense | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
General gene expression (GTEx)
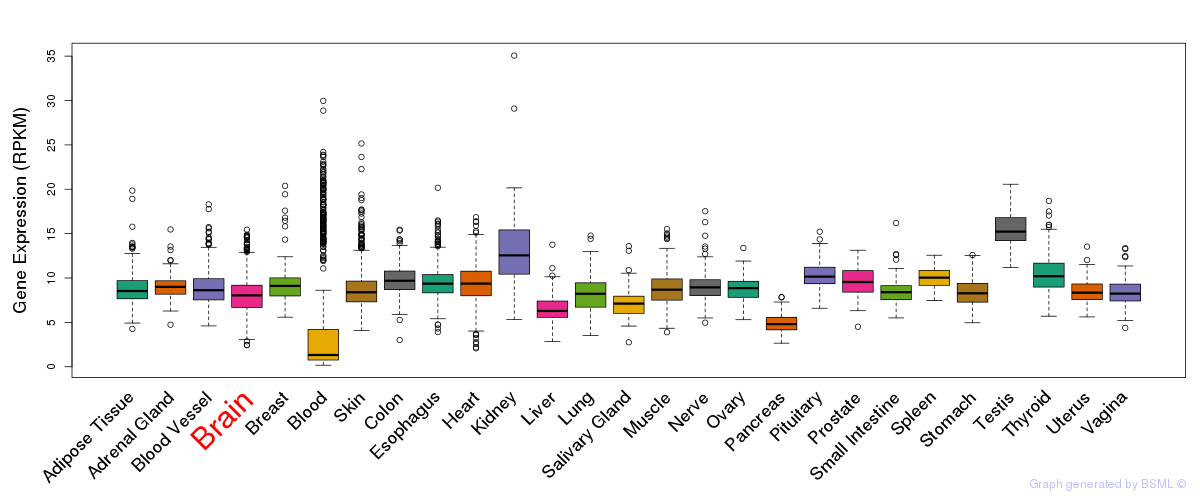
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003723 | RNA binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 15231747 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006397 | mRNA processing | TAS | 10369684 | |
| GO:0008380 | RNA splicing | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | TAS | 10369684 | |
| GO:0005681 | spliceosome | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| EEF1A1 | CCS-3 | CCS3 | EEF-1 | EEF1A | EF-Tu | EF1A | FLJ25721 | GRAF-1EF | HNGC:16303 | LENG7 | MGC102687 | MGC131894 | MGC16224 | PTI1 | eEF1A-1 | eukaryotic translation elongation factor 1 alpha 1 | Two-hybrid | BioGRID | 16169070 |
| GDF9 | - | growth differentiation factor 9 | Two-hybrid | BioGRID | 16169070 |
| LSM1 | CASM | YJL124C | LSM1 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD | 11920408 |
| LSM10 | MGC15749 | MST074 | MSTP074 | LSM10, U7 small nuclear RNA associated | Two-hybrid | BioGRID | 15231747 |16189514 |
| LSM2 | C6orf28 | G7b | YBL026W | snRNP | LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD | 11920408 |15231747 |
| LSM2 | C6orf28 | G7b | YBL026W | snRNP | LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) | Two-hybrid | BioGRID | 14667819 |15231747 |16169070 |16189514 |
| LSM4 | YER112W | LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD,BioGRID | 11920408 |
| LSM5 | FLJ12710 | YER146W | LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD | 11920408 |
| LSM6 | YDR378C | LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD | 11920408 |
| LSM7 | YNL147W | LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD,BioGRID | 11920408 |15231747 |
| LSM8 | YJR022W | LSM8 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD | 11920408 |
| SMN1 | BCD541 | SMA | SMA1 | SMA2 | SMA3 | SMA4 | SMA@ | SMN | SMNT | T-BCD541 | survival of motor neuron 1, telomeric | - | HPRD | 10851237 |
| SNRPD3 | SMD3 | small nuclear ribonucleoprotein D3 polypeptide 18kDa | - | HPRD,BioGRID | 15231747 |
| SNRPE | B-raf | SME | small nuclear ribonucleoprotein polypeptide E | - | HPRD,BioGRID | 15231747 |
| UTP14A | KIAA0266 | NY-CO-16 | SDCCAG16 | dJ537K23.3 | UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) | Two-hybrid | BioGRID | 16169070 |
| XPC | XP3 | XPCC | xeroderma pigmentosum, complementation group C | - | HPRD,BioGRID | 15231747 |
| XRN2 | - | 5'-3' exoribonuclease 2 | - | HPRD,BioGRID | 15231747 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG RNA DEGRADATION | 59 | 37 | All SZGR 2.0 genes in this pathway |
| KEGG SPLICEOSOME | 128 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | 15 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME DEADENYLATION DEPENDENT MRNA DECAY | 48 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| CHOW RASSF1 TARGETS UP | 27 | 17 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BASSO B LYMPHOCYTE NETWORK | 143 | 96 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| JIANG VHL TARGETS | 138 | 91 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 4 | 48 | 32 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE UP | 212 | 128 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 13 | 172 | 107 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA VIA VHL | 34 | 24 | All SZGR 2.0 genes in this pathway |
| LI DCP2 BOUND MRNA | 89 | 57 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 DN | 180 | 116 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN VINCRISTINE RESISTANCE B ALL DN | 15 | 10 | All SZGR 2.0 genes in this pathway |