Gene Page: ANK1
Summary ?
| GeneID | 286 |
| Symbol | ANK1 |
| Synonyms | ANK|SPH1|SPH2 |
| Description | ankyrin 1 |
| Reference | MIM:612641|HGNC:HGNC:492|Ensembl:ENSG00000029534|HPRD:01693|Vega:OTTHUMG00000150281 |
| Gene type | protein-coding |
| Map location | 8p11.1 |
| Pascal p-value | 0.416 |
| Fetal beta | -0.789 |
| DMG | 1 (# studies) |
| eGene | Meta |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| Expression | Meta-analysis of gene expression | P value: 1.772 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0437 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ANK1 | chr8 | 41571683 | G | C | NM_000037 NM_001142446 NM_020475 NM_020476 NM_020477 | p.597S>R p.630S>R p.597S>R p.597S>R p.597S>R | missense missense missense missense missense | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14904662 | 8 | 41655673 | ANK1 | 4.679E-4 | 0.533 | 0.046 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
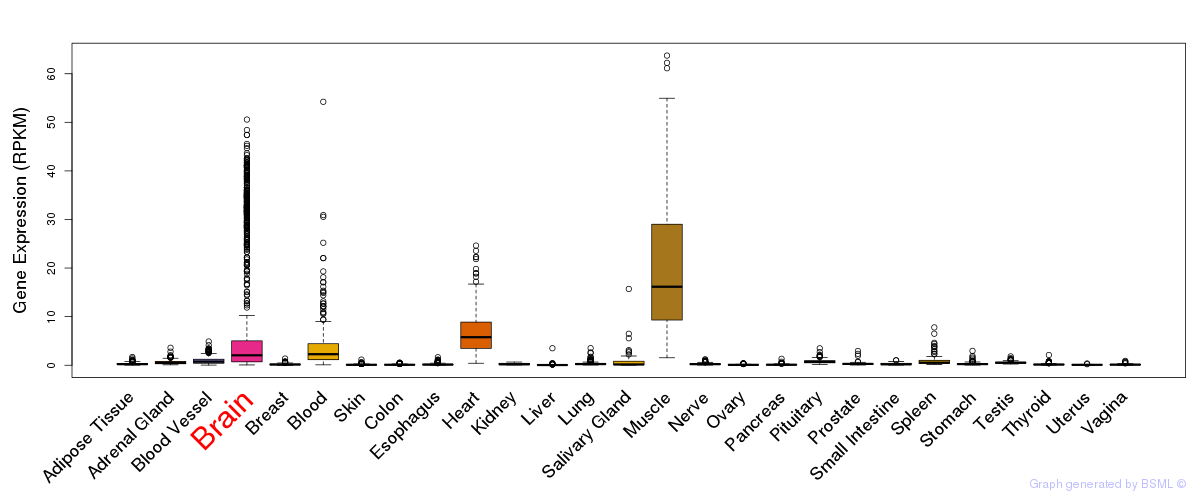
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005200 | structural constituent of cytoskeleton | TAS | 8640229 | |
| GO:0008093 | cytoskeletal adaptor activity | TAS | 11427698 | |
| GO:0030507 | spectrin binding | NAS | 8640229 | |
| GO:0019899 | enzyme binding | TAS | 11427698 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007010 | cytoskeleton organization | NAS | 9430667 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0006887 | exocytosis | NAS | 1833445 | |
| GO:0045199 | maintenance of epithelial cell apical/basal polarity | TAS | 11427698 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005856 | cytoskeleton | NAS | 1833445 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005886 | plasma membrane | NAS | 9430667 | |
| GO:0016323 | basolateral plasma membrane | NAS | 12409278 | |
| GO:0016529 | sarcoplasmic reticulum | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| L1CAM | CAML1 | CD171 | HSAS | HSAS1 | MASA | MIC5 | N-CAML1 | S10 | SPG1 | L1 cell adhesion molecule | - | HPRD,BioGRID | 11222639 |
| NBPF3 | AE2 | DKFZp434D177 | neuroblastoma breakpoint family, member 3 | - | HPRD | 9587054 |
| OBSCN | DKFZp666E245 | FLJ14124 | KIAA1556 | KIAA1639 | MGC120409 | MGC120410 | MGC120411 | MGC120412 | MGC138590 | UNC89 | obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF | - | HPRD,BioGRID | 12527750 |12631729 |
| RHAG | CD241 | RH2 | RH50A | Rh50 | Rh50GP | SLC42A1 | Rh-associated glycoprotein | - | HPRD,BioGRID | 12719424 |
| SLC4A1 | AE1 | BND3 | CD233 | DI | EMPB3 | EPB3 | FR | MGC116750 | MGC116753 | MGC126619 | MGC126623 | RTA1A | SW | WD | WD1 | WR | solute carrier family 4, anion exchanger, member 1 (erythrocyte membrane protein band 3, Diego blood group) | - | HPRD | 6449514 |
| SLC4A1 | AE1 | BND3 | CD233 | DI | EMPB3 | EPB3 | FR | MGC116750 | MGC116753 | MGC126619 | MGC126623 | RTA1A | SW | WD | WD1 | WR | solute carrier family 4, anion exchanger, member 1 (erythrocyte membrane protein band 3, Diego blood group) | Affinity Capture-Western | BioGRID | 8227202 |
| SLC4A3 | AE3 | SLC2C | solute carrier family 4, anion exchanger, member 3 | - | HPRD,BioGRID | 8227202 |
| SPTA1 | EL2 | SPTA | spectrin, alpha, erythrocytic 1 (elliptocytosis 2) | Reconstituted Complex | BioGRID | 2971657 |
| SPTAN1 | (ALPHA)II-SPECTRIN | FLJ44613 | NEAS | spectrin, alpha, non-erythrocytic 1 (alpha-fodrin) | - | HPRD | 492324 |2141335 |2970468 |2971657 |
| SPTB | HSpTB1 | spectrin, beta, erythrocytic | Reconstituted Complex | BioGRID | 2141335 |
| TIAM1 | FLJ36302 | T-cell lymphoma invasion and metastasis 1 | - | HPRD,BioGRID | 10893266 |
| TTN | CMD1G | CMH9 | CMPD4 | CONNECTIN | DKFZp451N061 | EOMFC | FLJ26020 | FLJ26409 | FLJ32040 | FLJ34413 | FLJ39564 | FLJ43066 | HMERF | LGMD2J | TMD | titin | - | HPRD,BioGRID | 12444090 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME L1CAM INTERACTIONS | 86 | 62 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | 23 | 19 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| DAVICIONI RHABDOMYOSARCOMA PAX FOXO1 FUSION UP | 64 | 37 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| KERLEY RESPONSE TO CISPLATIN UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION | 77 | 51 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8P12 P11 AMPLICON | 57 | 32 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY MUTATED AND AMPLIFIED IN BREAST CANCER | 94 | 60 | All SZGR 2.0 genes in this pathway |
| WELCH GATA1 TARGETS | 22 | 12 | All SZGR 2.0 genes in this pathway |
| ROSS AML OF FAB M7 TYPE | 68 | 44 | All SZGR 2.0 genes in this pathway |
| SCHURINGA STAT5A TARGETS UP | 21 | 13 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION DN | 184 | 132 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA UP | 52 | 33 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 7 | 28 | 16 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 8 | 26 | 17 | All SZGR 2.0 genes in this pathway |
| YOSHIOKA LIVER CANCER EARLY RECURRENCE UP | 40 | 23 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME2 | 17 | 11 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS DN | 242 | 146 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS DN | 187 | 109 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| STEINER ERYTHROCYTE MEMBRANE GENES | 15 | 10 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 2299 | 2306 | 1A,m8 | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-124.1 | 1482 | 1488 | 1A | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-128 | 631 | 637 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-138 | 899 | 906 | 1A,m8 | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| hsa-miR-138brain | AGCUGGUGUUGUGAAUC | ||||
| miR-139 | 137 | 144 | 1A,m8 | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
| miR-153 | 1015 | 1022 | 1A,m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-218 | 1526 | 1532 | 1A | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-27 | 631 | 637 | m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-328 | 1564 | 1570 | m8 | hsa-miR-328brain | CUGGCCCUCUCUGCCCUUCCGU |
| miR-448 | 1016 | 1022 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-452 | 1018 | 1024 | m8 | hsa-miR-452 | UGUUUGCAGAGGAAACUGAGAC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.