Gene Page: GRK5
Summary ?
| GeneID | 2869 |
| Symbol | GRK5 |
| Synonyms | GPRK5 |
| Description | G protein-coupled receptor kinase 5 |
| Reference | MIM:600870|HGNC:HGNC:4544|Ensembl:ENSG00000198873|HPRD:02926|Vega:OTTHUMG00000019149 |
| Gene type | protein-coding |
| Map location | 10q26.11 |
| Pascal p-value | 0.167 |
| Sherlock p-value | 0.133 |
| Fetal beta | 0.305 |
| DMG | 3 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09085932 | 10 | 121172083 | GRK5 | 4.422E-4 | 0.395 | 0.045 | DMG:Wockner_2014 |
| cg23303782 | 10 | 120967744 | GRK5 | -0.023 | 0.26 | DMG:Nishioka_2013 | |
| cg27315279 | 10 | 120966843 | GRK5 | 5.9E-9 | -0.013 | 3.17E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs3766034 | chr1 | 169087664 | GRK5 | 2869 | 0.05 | trans | ||
| rs6686962 | chr1 | 174953568 | GRK5 | 2869 | 0.04 | trans | ||
| rs10484468 | chr6 | 106083705 | GRK5 | 2869 | 0.17 | trans | ||
| rs11755501 | chr6 | 106084205 | GRK5 | 2869 | 0.17 | trans | ||
| rs6975312 | chr7 | 50620029 | GRK5 | 2869 | 0.13 | trans | ||
| rs11203077 | chr10 | 91097084 | GRK5 | 2869 | 0.04 | trans | ||
| rs17119672 | chr10 | 91100630 | GRK5 | 2869 | 0.01 | trans | ||
| rs7911396 | chr10 | 91101614 | GRK5 | 2869 | 0.01 | trans | ||
| rs7099668 | chr10 | 91107381 | GRK5 | 2869 | 0.01 | trans | ||
| rs7907341 | chr10 | 91108070 | GRK5 | 2869 | 0.01 | trans | ||
| rs12266581 | chr10 | 91109911 | GRK5 | 2869 | 0.01 | trans | ||
| rs1041540 | chr10 | 91111556 | GRK5 | 2869 | 0.01 | trans | ||
| rs4378299 | chr10 | 91111779 | GRK5 | 2869 | 0.01 | trans | ||
| rs6586191 | chr10 | 91135428 | GRK5 | 2869 | 0.01 | trans | ||
| rs12395234 | chrX | 52617968 | GRK5 | 2869 | 0.12 | trans | ||
| rs6568279 | 0 | GRK5 | 2869 | 0.16 | trans |
Section II. Transcriptome annotation
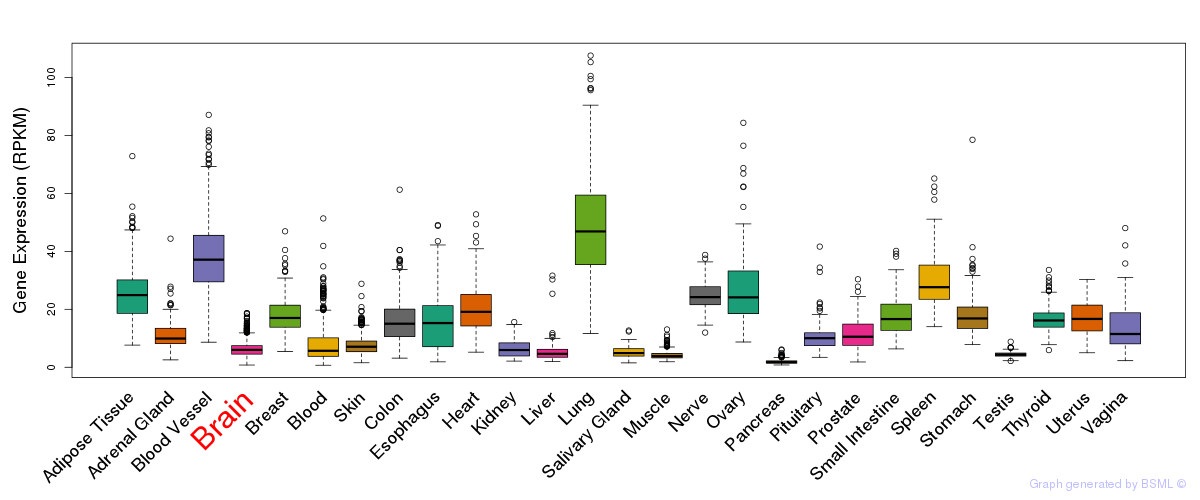
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FMNL1 | 0.56 | 0.53 |
| DDN | 0.56 | 0.50 |
| ICAM5 | 0.55 | 0.52 |
| PKD2L1 | 0.55 | 0.49 |
| CX3CL1 | 0.55 | 0.52 |
| KCNH3 | 0.55 | 0.55 |
| C2orf55 | 0.55 | 0.49 |
| ADAMTS8 | 0.54 | 0.48 |
| CHRM1 | 0.54 | 0.49 |
| FAM131A | 0.54 | 0.54 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SH2D2A | -0.31 | -0.36 |
| KIAA1949 | -0.31 | -0.22 |
| LEMD1 | -0.31 | -0.21 |
| AF186192.1 | -0.30 | -0.32 |
| TUBB2B | -0.30 | -0.26 |
| DYNLT1 | -0.29 | -0.33 |
| TIGD1 | -0.29 | -0.22 |
| AC005035.1 | -0.29 | -0.33 |
| TMSB15A | -0.29 | -0.24 |
| GPR125 | -0.29 | -0.30 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AGTR1 | AG2S | AGTR1A | AGTR1B | AT1 | AT1B | AT1R | AT2R1 | AT2R1A | AT2R1B | HAT1R | angiotensin II receptor, type 1 | Affinity Capture-Western | BioGRID | 9211925 |
| AVPR1A | AVPR1 | arginine vasopressin receptor 1A | - | HPRD,BioGRID | 10858434 |
| AVPR1B | AVPR3 | arginine vasopressin receptor 1B | - | HPRD | 10858434 |
| AVPR2 | ADHR | DI1 | DIR | DIR3 | MGC126533 | MGC138386 | NDI | V2R | arginine vasopressin receptor 2 | - | HPRD,BioGRID | 10858434 |
| CALM1 | CALML2 | CAMI | DD132 | PHKD | calmodulin 1 (phosphorylase kinase, delta) | Reconstituted Complex | BioGRID | 9092566 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | Reconstituted Complex | BioGRID | 10085129 |
| CHRM2 | FLJ43243 | HM2 | MGC120006 | MGC120007 | cholinergic receptor, muscarinic 2 | - | HPRD,BioGRID | 8288567 |
| GIT1 | - | G protein-coupled receptor kinase interacting ArfGAP 1 | - | HPRD | 9826657 |
| GRK5 | GPRK5 | G protein-coupled receptor kinase 5 | Biochemical Activity | BioGRID | 8144599 |
| OXTR | OT-R | oxytocin receptor | Affinity Capture-Western | BioGRID | 10858434 |
| RCVRN | RCV1 | recoverin | - | HPRD,BioGRID | 12507501 |
| RHO | CSNBAD1 | MGC138309 | MGC138311 | OPN2 | RP4 | rhodopsin | - | HPRD,BioGRID | 8144599 |8288567 |
| SNCA | MGC110988 | NACP | PARK1 | PARK4 | PD1 | synuclein, alpha (non A4 component of amyloid precursor) | Biochemical Activity | BioGRID | 10852916 |
| SNCB | - | synuclein, beta | Biochemical Activity | BioGRID | 10852916 |
| SNCG | BCSG1 | SR | synuclein, gamma (breast cancer-specific protein 1) | - | HPRD,BioGRID | 10852916 |
| TACR1 | NK1R | NKIR | SPR | TAC1R | tachykinin receptor 1 | - | HPRD,BioGRID | 12067742 |
| TBXA2R | TXA2-R | thromboxane A2 receptor | - | HPRD | 11504827 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| PID ALPHA SYNUCLEIN PATHWAY | 33 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | 205 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA Q SIGNALLING EVENTS | 184 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER BASAL VS MESENCHYMAL DN | 50 | 36 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL DN | 86 | 59 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY UP | 78 | 41 | All SZGR 2.0 genes in this pathway |
| PAPASPYRIDONOS UNSTABLE ATEROSCLEROTIC PLAQUE DN | 43 | 29 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS DN | 142 | 95 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 AND TP63 TARGETS | 205 | 145 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 XPCS DN | 88 | 71 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| XU HGF SIGNALING NOT VIA AKT1 48HR UP | 35 | 20 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| TOMLINS METASTASIS UP | 14 | 9 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN QTL CIS | 75 | 51 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS DN | 87 | 69 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP | 207 | 145 | All SZGR 2.0 genes in this pathway |
| YE METASTATIC LIVER CANCER | 27 | 13 | All SZGR 2.0 genes in this pathway |
| ZHANG TARGETS OF EWSR1 FLI1 FUSION | 88 | 68 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR UP | 78 | 56 | All SZGR 2.0 genes in this pathway |
| STOSSI RESPONSE TO ESTRADIOL | 50 | 35 | All SZGR 2.0 genes in this pathway |
| HESS TARGETS OF HOXA9 AND MEIS1 DN | 77 | 48 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| MA PITUITARY FETAL VS ADULT DN | 19 | 17 | All SZGR 2.0 genes in this pathway |
| XU GH1 EXOGENOUS TARGETS UP | 85 | 50 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| OUILLETTE CLL 13Q14 DELETION DN | 60 | 38 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS MODEL UP | 45 | 26 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 13 | 30 | 20 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS UP | 91 | 59 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN UP | 90 | 58 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION UP | 271 | 165 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |