Gene Page: GRIN1
Summary ?
| GeneID | 2902 |
| Symbol | GRIN1 |
| Synonyms | GluN1|MRD8|NMD-R1|NMDA1|NMDAR1|NR1 |
| Description | glutamate ionotropic receptor NMDA type subunit 1 |
| Reference | MIM:138249|HGNC:HGNC:4584|Ensembl:ENSG00000176884|HPRD:15926|Vega:OTTHUMG00000020976 |
| Gene type | protein-coding |
| Map location | 9q34.3 |
| Pascal p-value | 0.108 |
| Sherlock p-value | 0.397 |
| Fetal beta | -1.27 |
| eGene | Myers' cis & trans |
| Support | LIGAND GATED ION SIGNALING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_PocklingtonH1 G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanNRC CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 15 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 7.9864 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs3823720 | chr7 | 42001170 | GRIN1 | 2902 | 0.19 | trans |
Section II. Transcriptome annotation
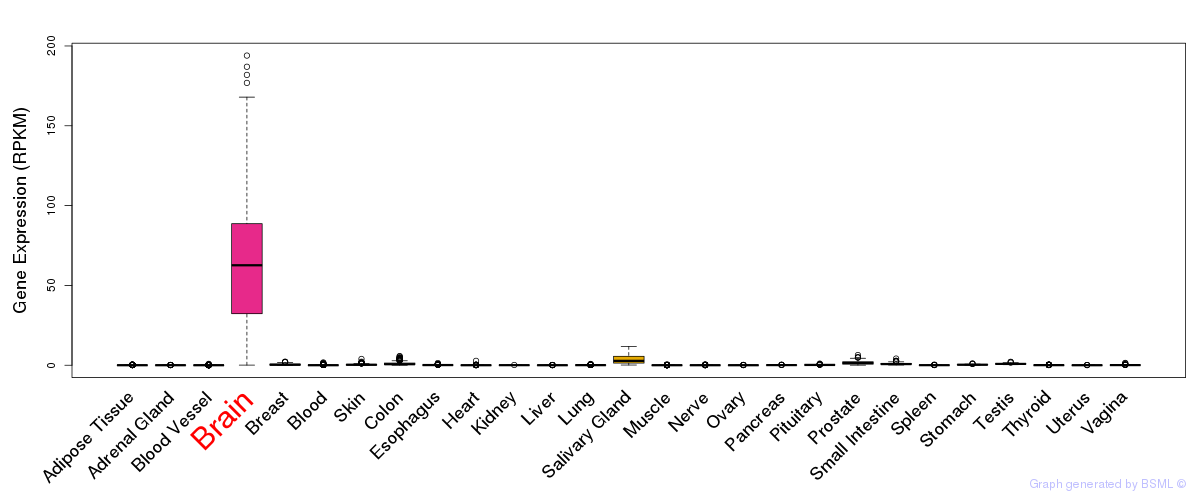
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC021914.1 | 0.74 | 0.28 |
| CLEC2B | 0.64 | 0.35 |
| CXCL2 | 0.61 | 0.53 |
| AL050337.1 | 0.57 | 0.18 |
| AC137766.1 | 0.56 | 0.37 |
| C1orf61 | 0.55 | 0.34 |
| C17orf84 | 0.54 | 0.03 |
| OASL | 0.53 | 0.33 |
| C1orf54 | 0.52 | 0.31 |
| IFITM1 | 0.51 | 0.47 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DNAJC7 | -0.51 | -0.38 |
| CHMP4B | -0.50 | -0.35 |
| ARFGAP2 | -0.50 | -0.40 |
| NUDC | -0.49 | -0.40 |
| NDUFS2 | -0.48 | -0.36 |
| UBE2Q1 | -0.48 | -0.37 |
| GRPEL1 | -0.48 | -0.40 |
| C20orf43 | -0.47 | -0.36 |
| SPG7 | -0.47 | -0.29 |
| MRPS5 | -0.47 | -0.41 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004972 | N-methyl-D-aspartate selective glutamate receptor activity | IDA | glutamate (GO term level: 8) | 7679115 |7685113 |
| GO:0004970 | ionotropic glutamate receptor activity | IEA | glutamate (GO term level: 7) | - |
| GO:0005234 | extracellular-glutamate-gated ion channel activity | IEA | glutamate (GO term level: 11) | - |
| GO:0016595 | glutamate binding | IDA | glutamate (GO term level: 5) | 7685113 |
| GO:0035254 | glutamate receptor binding | ISS | glutamate (GO term level: 5) | - |
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005509 | calcium ion binding | ISS | - | |
| GO:0005516 | calmodulin binding | ISS | - | |
| GO:0005262 | calcium channel activity | IDA | 7685113 | |
| GO:0005216 | ion channel activity | IEA | - | |
| GO:0016594 | glycine binding | IDA | 7685113 | |
| GO:0016594 | glycine binding | IMP | 17047094 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0060079 | regulation of excitatory postsynaptic membrane potential | ISS | neuron, Synap (GO term level: 10) | - |
| GO:0007268 | synaptic transmission | ISS | neuron, Synap, Neurotransmitter (GO term level: 6) | - |
| GO:0035235 | ionotropic glutamate receptor signaling pathway | ISS | glutamate (GO term level: 8) | - |
| GO:0008542 | visual learning | ISS | - | |
| GO:0006811 | ion transport | IEA | - | |
| GO:0006812 | cation transport | IDA | 7685113 | |
| GO:0042391 | regulation of membrane potential | IDA | 7679115 |7685113 |17047094 | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | ISS | - | |
| GO:0045471 | response to ethanol | IDA | 18445116 | |
| GO:0055074 | calcium ion homeostasis | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008021 | synaptic vesicle | ISS | Synap, Neurotransmitter (GO term level: 12) | - |
| GO:0014069 | postsynaptic density | ISS | Synap (GO term level: 10) | - |
| GO:0019717 | synaptosome | ISS | Synap, Brain (GO term level: 7) | - |
| GO:0017146 | N-methyl-D-aspartate selective glutamate receptor complex | IDA | glutamate, Synap (GO term level: 10) | 7679115 |10480938 |17047094 |
| GO:0030425 | dendrite | IDA | neuron, axon, dendrite (GO term level: 6) | 10749211 |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045211 | postsynaptic membrane | ISS | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0045202 | synapse | ISS | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | IDA | 7685113 | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTN2 | CMD1AA | actinin, alpha 2 | - | HPRD | 9009191 |9728925 |9952395 |
| ACTN2 | CMD1AA | actinin, alpha 2 | alpha-actinin-2 interacts with NR1A. This interaction was modeled on a demonstrated interaction between human alpha-actinin-2 and NR1A from an unspecified species. | BIND | 9009191 |
| AKAP9 | AKAP350 | AKAP450 | CG-NAP | HYPERION | KIAA0803 | MU-RMS-40.16A | PRKA9 | YOTIAO | A kinase (PRKA) anchor protein (yotiao) 9 | - | HPRD | 9482789|10862698 |
| AKAP9 | AKAP350 | AKAP450 | CG-NAP | HYPERION | KIAA0803 | MU-RMS-40.16A | PRKA9 | YOTIAO | A kinase (PRKA) anchor protein (yotiao) 9 | - | HPRD | 10862698 |
| CACNG2 | MGC138502 | MGC138504 | calcium channel, voltage-dependent, gamma subunit 2 | Co-fractionation | BioGRID | 11140673 |
| CAMK2A | CAMKA | KIAA0968 | calcium/calmodulin-dependent protein kinase II alpha | - | HPRD | 10862698 |
| CANX | CNX | FLJ26570 | IP90 | P90 | calnexin | - | HPRD | 11506858 |14732708 |
| CDH2 | CD325 | CDHN | CDw325 | NCAD | cadherin 2, type 1, N-cadherin (neuronal) | - | HPRD | 10862698 |
| CIT | CRIK | KIAA0949 | STK21 | citron (rho-interacting, serine/threonine kinase 21) | - | HPRD | 10862698 |
| CLTC | CHC | CHC17 | CLH-17 | CLTCL2 | Hc | KIAA0034 | clathrin, heavy chain (Hc) | - | HPRD | 10862698 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | - | HPRD | 10862698 |
| DLG2 | DKFZp781D1854 | DKFZp781E0954 | FLJ37266 | MGC131811 | PSD-93 | discs, large homolog 2, chapsyn-110 (Drosophila) | - | HPRD | 10862698 |
| DLG3 | KIAA1232 | MRX | MRX90 | NE-Dlg | NEDLG | SAP102 | discs, large homolog 3 (neuroendocrine-dlg, Drosophila) | - | HPRD | 10862698 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | - | HPRD | 9009191 |9502803 |11506858 |12068077 |14732708 |15030493 |
| DLGAP4 | DAP4 | KIAA0964 | MGC131862 | RP5-977B1.6 | SAPAP4 | discs, large (Drosophila) homolog-associated protein 4 | - | HPRD | 10862698 |
| DNM1 | DNM | dynamin 1 | - | HPRD | 10862698 |
| DRD1 | DADR | DRD1A | dopamine receptor D1 | - | HPRD | 12408866 |
| DUSP4 | HVH2 | MKP-2 | MKP2 | TYP | dual specificity phosphatase 4 | - | HPRD | 10862698 |
| EPHB2 | CAPB | DRT | EPHT3 | ERK | Hek5 | MGC87492 | PCBC | Tyro5 | EPH receptor B2 | - | HPRD | 11754835 |
| EPHB4 | HTK | MYK1 | TYRO11 | EPH receptor B4 | - | HPRD | 11136979 |
| FUS | CHOP | FUS-CHOP | FUS1 | TLS | TLS/CHOP | hnRNP-P2 | fusion (involved in t(12;16) in malignant liposarcoma) | - | HPRD | 10862698 |
| GRIN2A | NMDAR2A | NR2A | glutamate receptor, ionotropic, N-methyl D-aspartate 2A | NR1a interacts with NR2A. | BIND | 11279200 |
| GRIN2A | NMDAR2A | NR2A | glutamate receptor, ionotropic, N-methyl D-aspartate 2A | - | HPRD | 8139656 |10862698 |11803122 |12068077 |12191494 |12893641 |14732708 |
| GRIN2A | NMDAR2A | NR2A | glutamate receptor, ionotropic, N-methyl D-aspartate 2A | Affinity Capture-Western | BioGRID | 12391275 |
| GRIN2B | MGC142178 | MGC142180 | NMDAR2B | NR2B | hNR3 | glutamate receptor, ionotropic, N-methyl D-aspartate 2B | Affinity Capture-Western | BioGRID | 12391275 |
| GRIN2B | MGC142178 | MGC142180 | NMDAR2B | NR2B | hNR3 | glutamate receptor, ionotropic, N-methyl D-aspartate 2B | - | HPRD | 8139656 |12191494 |15030493 |
| GRIN2D | EB11 | NMDAR2D | glutamate receptor, ionotropic, N-methyl D-aspartate 2D | - | HPRD | 12191494 |
| GRIN3A | FLJ45414 | NMDAR-L | NR3A | glutamate receptor, ionotropic, N-methyl-D-aspartate 3A | Affinity Capture-Western | BioGRID | 12391275 |
| GRIN3A | FLJ45414 | NMDAR-L | NR3A | glutamate receptor, ionotropic, N-methyl-D-aspartate 3A | - | HPRD | 9620802 |11160393 |11588171 |
| GRIN3B | NR3B | glutamate receptor, ionotropic, N-methyl-D-aspartate 3B | Affinity Capture-Western | BioGRID | 12008020 |
| HNRNPU | HNRPU | SAF-A | U21.1 | heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) | - | HPRD | 10862698 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | - | HPRD | 10862698 |
| HSPA1A | FLJ54303 | FLJ54370 | FLJ54392 | FLJ54408 | FLJ75127 | HSP70-1 | HSP70-1A | HSP70I | HSP72 | HSPA1 | HSPA1B | heat shock 70kDa protein 1A | - | HPRD | 10862698 |
| INA | FLJ18662 | FLJ57501 | MGC12702 | NEF5 | NF-66 | TXBP-1 | internexin neuronal intermediate filament protein, alpha | - | HPRD | 10862698 |
| KAT5 | ESA1 | HTATIP | HTATIP1 | PLIP | TIP | TIP60 | cPLA2 | K(lysine) acetyltransferase 5 | - | HPRD | 10862698 |
| MAP2 | DKFZp686I2148 | MAP2A | MAP2B | MAP2C | microtubule-associated protein 2 | - | HPRD | 10862698 |
| MAP2K2 | FLJ26075 | MAPKK2 | MEK2 | MKK2 | PRKMK2 | mitogen-activated protein kinase kinase 2 | - | HPRD | 10862698 |
| MYH9 | DFNA17 | EPSTS | FTNS | MGC104539 | MHA | NMHC-II-A | NMMHCA | myosin, heavy chain 9, non-muscle | - | HPRD | 10862698 |
| NANOS1 | NOS1 | nanos homolog 1 (Drosophila) | - | HPRD | 10862698 |
| ND2 | MTND2 | NADH dehydrogenase, subunit 2 (complex I) | Affinity Capture-Western | BioGRID | 15069201 |
| NEFL | CMT1F | CMT2E | FLJ53642 | NF-L | NF68 | NFL | neurofilament, light polypeptide | - | HPRD | 9425014 |
| NF1 | DKFZp686J1293 | FLJ21220 | NFNS | VRNF | WSS | neurofibromin 1 | - | HPRD | 10862698 |
| PLAT | DKFZp686I03148 | T-PA | TPA | plasminogen activator, tissue | - | HPRD | 15448144 |
| PPP2R1A | MGC786 | PR65A | protein phosphatase 2 (formerly 2A), regulatory subunit A, alpha isoform | - | HPRD | 10862698 |
| PPP2R2A | B55-ALPHA | B55A | FLJ26613 | MGC52248 | PR52A | PR55A | protein phosphatase 2 (formerly 2A), regulatory subunit B, alpha isoform | - | HPRD | 10862698 |
| PRKCB | MGC41878 | PKC-beta | PKCB | PRKCB1 | PRKCB2 | protein kinase C, beta | - | HPRD | 10862698 |
| PRKCE | MGC125656 | MGC125657 | PKCE | nPKC-epsilon | protein kinase C, epsilon | - | HPRD | 10862698 |
| PRKCG | MGC57564 | PKC-gamma | PKCC | PKCG | SCA14 | protein kinase C, gamma | - | HPRD | 10862698 |
| RAP2A | K-REV | KREV | RAP2 | RbBP-30 | RAP2A, member of RAS oncogene family | - | HPRD | 10862698 |
| RPS6KA3 | CLS | HU-3 | ISPK-1 | MAPKAPK1B | MRX19 | RSK | RSK2 | S6K-alpha3 | p90-RSK2 | pp90RSK2 | ribosomal protein S6 kinase, 90kDa, polypeptide 3 | - | HPRD | 10862698 |
| SP1 | - | Sp1 transcription factor | - | HPRD | 14970236 |
| SP3 | DKFZp686O1631 | SPR-2 | Sp3 transcription factor | - | HPRD | 14970236 |
| SPTAN1 | (ALPHA)II-SPECTRIN | FLJ44613 | NEAS | spectrin, alpha, non-erythrocytic 1 (alpha-fodrin) | - | HPRD | 9670010 |10862698 |
| SYNGAP1 | DKFZp761G1421 | KIAA1938 | RASA1 | RASA5 | SYNGAP | synaptic Ras GTPase activating protein 1 homolog (rat) | - | HPRD | 10862698 |
| TIAM1 | FLJ36302 | T-cell lymphoma invasion and metastasis 1 | Tiam1 interacts with NMDA subunit NR1. This interaction was modelled on a demonstrated interaction between human Tiam1 and NR1 from an unspecified species. | BIND | 15721239 |
| TJP1 | DKFZp686M05161 | MGC133289 | ZO-1 | tight junction protein 1 (zona occludens 1) | - | HPRD | 10862698 |
| TRAF3 | CAP-1 | CD40bp | CRAF1 | LAP1 | TNF receptor-associated factor 3 | - | HPRD | 10862698 |
| TUBA4B | FLJ13940 | TUBA4 | tubulin, alpha 4b (pseudogene) | - | HPRD | 10862698 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM POTENTIATION | 70 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| KEGG AMYOTROPHIC LATERAL SCLEROSIS ALS | 53 | 43 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EPONFKB PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NOS1 PATHWAY | 24 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | 17 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | 37 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | 33 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | 15 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | 15 | 14 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL DN | 186 | 107 | All SZGR 2.0 genes in this pathway |
| EBAUER TARGETS OF PAX3 FOXO1 FUSION UP | 207 | 128 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| CHIARETTI ACUTE LYMPHOBLASTIC LEUKEMIA ZAP70 | 67 | 33 | All SZGR 2.0 genes in this pathway |
| MCCLUNG DELTA FOSB TARGETS 8WK | 47 | 38 | All SZGR 2.0 genes in this pathway |
| ULE SPLICING VIA NOVA2 | 43 | 38 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN | 88 | 58 | All SZGR 2.0 genes in this pathway |
| MARIADASON RESPONSE TO BUTYRATE SULINDAC 6 | 52 | 32 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN UP | 181 | 112 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-15/16/195/424/497 | 58 | 64 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-214 | 56 | 62 | m8 | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.