Gene Page: GSN
Summary ?
| GeneID | 2934 |
| Symbol | GSN |
| Synonyms | ADF|AGEL |
| Description | gelsolin |
| Reference | MIM:137350|HGNC:HGNC:4620|Ensembl:ENSG00000148180|HPRD:00674|Vega:OTTHUMG00000020584 |
| Gene type | protein-coding |
| Map location | 9q33 |
| Pascal p-value | 0.064 |
| Sherlock p-value | 0.187 |
| Fetal beta | -2.469 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.humanNRC CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg13584244 | 9 | 124030048 | GSN | 7.65E-5 | -0.578 | 0.025 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs798365 | chr2 | 16896755 | GSN | 2934 | 0.15 | trans | ||
| rs317971 | chr5 | 66688724 | GSN | 2934 | 0.18 | trans | ||
| rs6934246 | chr6 | 35154494 | GSN | 2934 | 0.19 | trans | ||
| rs17792062 | chr8 | 129163736 | GSN | 2934 | 0.09 | trans | ||
| rs6482363 | chr10 | 24368607 | GSN | 2934 | 0.07 | trans | ||
| rs11049036 | chr12 | 27650292 | GSN | 2934 | 0.1 | trans | ||
| rs10879027 | chr12 | 70237156 | GSN | 2934 | 0.12 | trans |
Section II. Transcriptome annotation
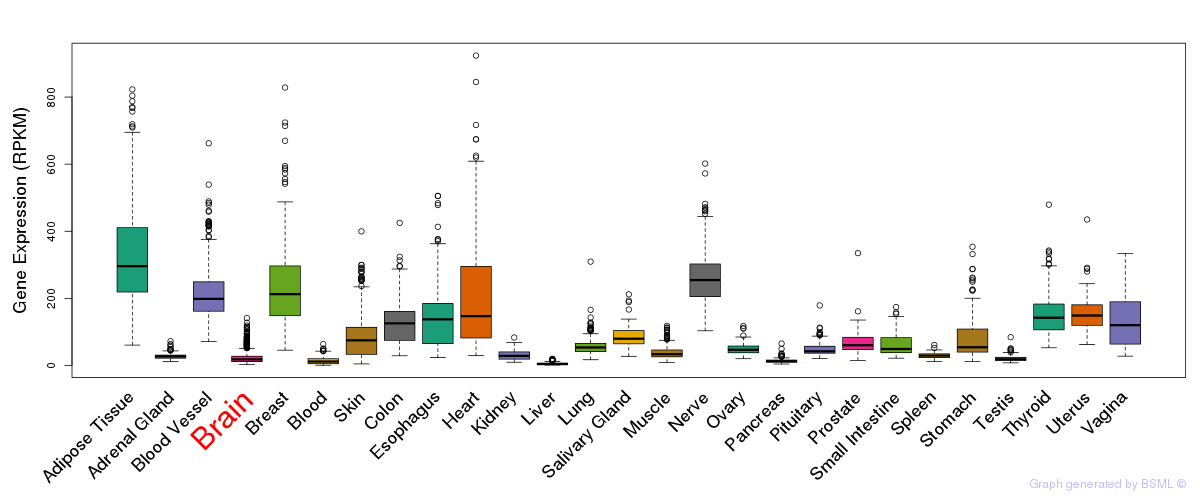
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTA1 | ACTA | ASMA | CFTD | CFTD1 | CFTDM | MPFD | NEM1 | NEM2 | NEM3 | actin, alpha 1, skeletal muscle | - | HPRD,BioGRID | 10583954 |
| ACTB | PS1TP5BP1 | actin, beta | - | HPRD | 10945978 |
| ACTN4 | ACTININ-4 | DKFZp686K23158 | FSGS | FSGS1 | actinin, alpha 4 | - | HPRD,BioGRID | 11733011 |
| APP | AAA | ABETA | ABPP | AD1 | APPI | CTFgamma | CVAP | PN2 | amyloid beta (A4) precursor protein | - | HPRD,BioGRID | 10329371 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD,BioGRID | 12941811 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | - | HPRD | 11577104 |
| CASP4 | ICE(rel)II | ICEREL-II | ICH-2 | Mih1/TX | TX | caspase 4, apoptosis-related cysteine peptidase | Affinity Capture-MS | BioGRID | 17353931 |
| CLIC5 | CLIC5B | FLJ90663 | MST130 | MSTP130 | dJ447E21.4 | chloride intracellular channel 5 | - | HPRD | 10793131 |
| FN1 | CIG | DKFZp686F10164 | DKFZp686H0342 | DKFZp686I1370 | DKFZp686O13149 | ED-B | FINC | FN | FNZ | GFND | GFND2 | LETS | MSF | fibronectin 1 | - | HPRD,BioGRID | 6092370 |
| LIMK2 | - | LIM domain kinase 2 | - | HPRD | 11171090 |
| PIK3CA | MGC142161 | MGC142163 | PI3K | p110-alpha | phosphoinositide-3-kinase, catalytic, alpha polypeptide | Affinity Capture-Western | BioGRID | 11577104 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | Reconstituted Complex | BioGRID | 9164868 |
| PPM1B | MGC21657 | PP2C-beta-X | PP2CB | PP2CBETA | PPC2BETAX | protein phosphatase 1B (formerly 2C), magnesium-dependent, beta isoform | Affinity Capture-MS | BioGRID | 17353931 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | Affinity Capture-Western | BioGRID | 11577104 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD,BioGRID | 12578912 |
| PTPN12 | PTP-PEST | PTPG1 | protein tyrosine phosphatase, non-receptor type 12 | Affinity Capture-Western | BioGRID | 11577104 |
| PXN | FLJ16691 | paxillin | - | HPRD,BioGRID | 11577104 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Affinity Capture-Western | BioGRID | 11577104 |
| STK39 | DCHT | DKFZp686K05124 | PASK | SPAK | serine threonine kinase 39 (STE20/SPS1 homolog, yeast) | Two-hybrid | BioGRID | 14563843 |
| TNIK | - | TRAF2 and NCK interacting kinase | - | HPRD | 10521462 |
| TOM1L1 | OK/KNS-CL.3 | SRCASM | target of myb1 (chicken)-like 1 | Two-hybrid | BioGRID | 16169070 |
| VASP | - | vasodilator-stimulated phosphoprotein | - | HPRD | 11093254 |
| VCL | CMD1W | MVCL | vinculin | - | HPRD,BioGRID | 11577104 |
| VDAC1 | MGC111064 | PORIN | PORIN-31-HL | voltage-dependent anion channel 1 | - | HPRD,BioGRID | 11039896 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FC GAMMA R MEDIATED PHAGOCYTOSIS | 97 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ECM PATHWAY | 24 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIVNEF PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RHO PATHWAY | 32 | 23 | All SZGR 2.0 genes in this pathway |
| PID AR PATHWAY | 61 | 46 | All SZGR 2.0 genes in this pathway |
| PID AVB3 OPN PATHWAY | 31 | 29 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 RECEPTOR PROXIMAL PATHWAY | 35 | 29 | All SZGR 2.0 genes in this pathway |
| PID CASPASE PATHWAY | 52 | 39 | All SZGR 2.0 genes in this pathway |
| PID NCADHERIN PATHWAY | 33 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | 40 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME AMYLOIDS | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC EXECUTION PHASE | 54 | 37 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT EARLY UP | 21 | 19 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER BASAL VS MESENCHYMAL UP | 121 | 45 | All SZGR 2.0 genes in this pathway |
| RODRIGUES DCC TARGETS DN | 121 | 84 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP | 185 | 126 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL UP | 146 | 99 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS DN | 142 | 95 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2A DN | 141 | 84 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| MISSIAGLIA REGULATED BY METHYLATION UP | 126 | 78 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK UP | 214 | 144 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS UP | 169 | 127 | All SZGR 2.0 genes in this pathway |
| MCBRYAN TERMINAL END BUD DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| MARKS HDAC TARGETS UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| TOMLINS PROSTATE CANCER DN | 40 | 33 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE UP | 77 | 60 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| MORI IMMATURE B LYMPHOCYTE DN | 90 | 55 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS DN | 87 | 69 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT UP | 165 | 100 | All SZGR 2.0 genes in this pathway |
| HASLINGER B CLL WITH 11Q23 DELETION | 23 | 18 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION DN | 184 | 132 | All SZGR 2.0 genes in this pathway |
| DORSEY GAB2 TARGETS | 31 | 17 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| XU CREBBP TARGETS DN | 44 | 31 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS UP | 151 | 103 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS DN | 74 | 55 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| GAJATE RESPONSE TO TRABECTEDIN UP | 67 | 45 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C7 | 68 | 44 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| LEIN ASTROCYTE MARKERS | 42 | 35 | All SZGR 2.0 genes in this pathway |
| LEIN OLIGODENDROCYTE MARKERS | 74 | 53 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| ENGELMANN CANCER PROGENITORS DN | 70 | 44 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A UP | 111 | 70 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA D CLUSTER DN | 40 | 26 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA D DN | 78 | 34 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS UP | 153 | 107 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN | 209 | 137 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE UP | 163 | 102 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS UP | 108 | 78 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR UP | 101 | 65 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA PCA3 DN | 69 | 38 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3 UNMETHYLATED | 228 | 119 | All SZGR 2.0 genes in this pathway |
| BAE BRCA1 TARGETS UP | 75 | 47 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS DN | 207 | 139 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 UP | 134 | 85 | All SZGR 2.0 genes in this pathway |
| OHGUCHI LIVER HNF4A TARGETS UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| ISSAEVA MLL2 TARGETS | 62 | 35 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| DELPUECH FOXO3 TARGETS UP | 68 | 49 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP | 194 | 133 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS UP | 149 | 96 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |