Gene Page: HDAC1
Summary ?
| GeneID | 3065 |
| Symbol | HDAC1 |
| Synonyms | GON-10|HD1|RPD3|RPD3L1 |
| Description | histone deacetylase 1 |
| Reference | MIM:601241|HGNC:HGNC:4852|Ensembl:ENSG00000116478|HPRD:03143|Vega:OTTHUMG00000007529 |
| Gene type | protein-coding |
| Map location | 1p34 |
| Pascal p-value | 0.59 |
| Fetal beta | 0.69 |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4501739 | chrX | 35960848 | HDAC1 | 3065 | 0.03 | trans |
Section II. Transcriptome annotation
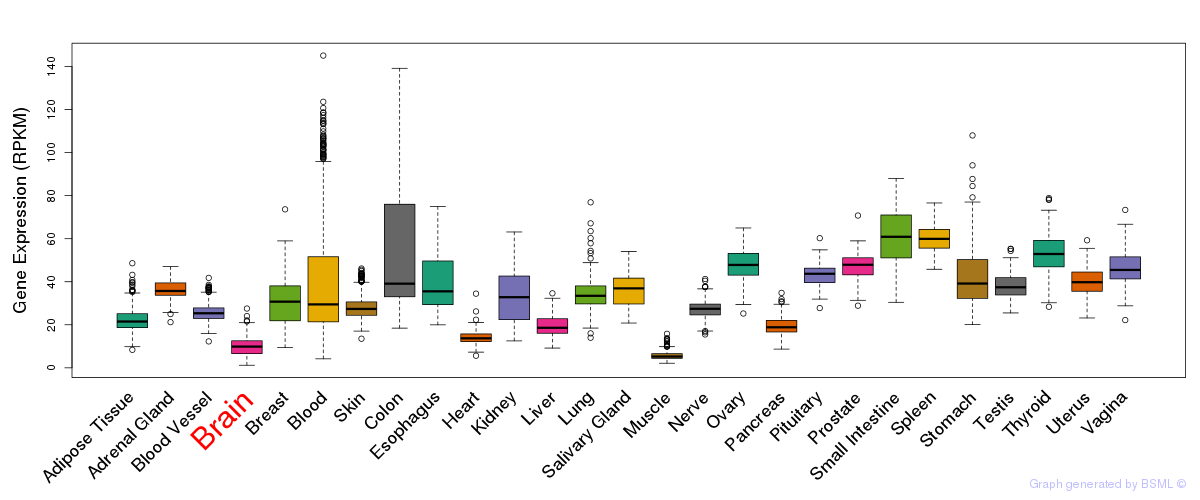
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MAMDC4 | 0.90 | 0.78 |
| AC135050.2 | 0.89 | 0.76 |
| CNKSR1 | 0.89 | 0.78 |
| ZNF692 | 0.89 | 0.81 |
| VWCE | 0.89 | 0.77 |
| MST1 | 0.88 | 0.77 |
| MYO19 | 0.88 | 0.76 |
| VARS2 | 0.88 | 0.70 |
| CTRL | 0.88 | 0.78 |
| GAL3ST4 | 0.88 | 0.67 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ABCG2 | -0.51 | -0.64 |
| HLA-C | -0.49 | -0.51 |
| FBXO2 | -0.47 | -0.50 |
| PLA2G4A | -0.46 | -0.58 |
| ITM2B | -0.46 | -0.53 |
| AIFM3 | -0.45 | -0.46 |
| ALDOC | -0.45 | -0.45 |
| RGS5 | -0.45 | -0.57 |
| SLC39A8 | -0.45 | -0.54 |
| HLA-F | -0.45 | -0.40 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | TAS | 8646880 | |
| GO:0004407 | histone deacetylase activity | TAS | 12711221 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0008134 | transcription factor binding | IPI | 15509593 | |
| GO:0008134 | transcription factor binding | TAS | 12711221 | |
| GO:0042802 | identical protein binding | IPI | 9150135 | |
| GO:0019899 | enzyme binding | IPI | 11062478 |11136718 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006916 | anti-apoptosis | TAS | 10777477 | |
| GO:0016568 | chromatin modification | TAS | 12711221 | |
| GO:0016575 | histone deacetylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000118 | histone deacetylase complex | TAS | 12711221 | |
| GO:0005634 | nucleus | IDA | 10846170 | |
| GO:0005634 | nucleus | TAS | 12711221 | |
| GO:0005737 | cytoplasm | TAS | 12711221 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AOF2 | BHC110 | KDM1 | KIAA0601 | LSD1 | amine oxidase (flavin containing) domain 2 | - | HPRD,BioGRID | 12493763 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD,BioGRID | 11994312 |
| ARID4A | RBBP1 | RBP-1 | RBP1 | AT rich interactive domain 4A (RBP1-like) | - | HPRD | 10490602 |
| ARID4B | BCAA | BRCAA1 | DKFZp313M2420 | MGC163290 | RBBP1L1 | RBP1L1 | SAP180 | AT rich interactive domain 4B (RBP1-like) | Affinity Capture-Western | BioGRID | 12724404 |
| ATR | FRP1 | MEC1 | SCKL | SCKL1 | ataxia telangiectasia and Rad3 related | Affinity Capture-Western | BioGRID | 10545197 |
| ATRX | ATR2 | MGC2094 | MRXHF1 | RAD54 | RAD54L | SFM1 | SHS | XH2 | XNP | ZNF-HX | alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) | - | HPRD | 14645126 |
| BAZ2A | DKFZp781B109 | FLJ13768 | FLJ13780 | FLJ45876 | KIAA0314 | TIP5 | WALp3 | bromodomain adjacent to zinc finger domain, 2A | - | HPRD | 12198165 |
| BCL3 | BCL4 | D19S37 | B-cell CLL/lymphoma 3 | HDAC1 interacts with BCL-3. | BIND | 15469820 |
| BCL6 | BCL5 | BCL6A | LAZ3 | ZBTB27 | ZNF51 | B-cell CLL/lymphoma 6 | Affinity Capture-Western | BioGRID | 9627120 |
| BCOR | ANOP2 | FLJ20285 | FLJ38041 | KIAA1575 | MAA2 | MCOPS2 | MGC131961 | MGC71031 | BCL6 co-repressor | - | HPRD,BioGRID | 10898795 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | BRCA1 interacts with HDAC1 | BIND | 10220405 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD,BioGRID | 10220405 |
| BUB1 | BUB1A | BUB1L | hBUB1 | budding uninhibited by benzimidazoles 1 homolog (yeast) | Affinity Capture-Western Co-purification | BioGRID | 15388328 |
| BUB1B | BUB1beta | BUBR1 | Bub1A | MAD3L | SSK1 | hBUBR1 | budding uninhibited by benzimidazoles 1 homolog beta (yeast) | Affinity Capture-Western Co-purification | BioGRID | 15388328 |
| BUB3 | BUB3L | hBUB3 | budding uninhibited by benzimidazoles 3 homolog (yeast) | Affinity Capture-Western Co-purification | BioGRID | 15388328 |
| CABIN1 | CAIN | KIAA0330 | PPP3IN | calcineurin binding protein 1 | - | HPRD,BioGRID | 10933397 |
| CBFA2T3 | ETO2 | MTG16 | MTGR2 | ZMYND4 | core-binding factor, runt domain, alpha subunit 2; translocated to, 3 | - | HPRD,BioGRID | 12242670 |
| CBX5 | HP1 | HP1A | chromobox homolog 5 (HP1 alpha homolog, Drosophila) | Affinity Capture-Western | BioGRID | 12242305 |
| CDC20 | CDC20A | MGC102824 | bA276H19.3 | p55CDC | cell division cycle 20 homolog (S. cerevisiae) | Affinity Capture-Western Co-purification | BioGRID | 15388328 |
| CDH1 | Arc-1 | CD324 | CDHE | ECAD | LCAM | UVO | cadherin 1, type 1, E-cadherin (epithelial) | Affinity Capture-Western Co-purification | BioGRID | 15388328 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | HDAC1 interacts with the p21 promoter. | BIND | 15710329 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | HDAC1 interacts with the CDKN1A (p21) promoter. | BIND | 15674334 |
| CDYL | CDYL1 | DKFZp586C1622 | MGC131936 | chromodomain protein, Y-like | - | HPRD,BioGRID | 12947414 |
| CHD1 | DKFZp686E2337 | chromodomain helicase DNA binding protein 1 | - | HPRD,BioGRID | 12890497 |
| CHD3 | Mi-2a | Mi2-ALPHA | ZFH | chromodomain helicase DNA binding protein 3 | - | HPRD,BioGRID | 9804427 |
| CHD4 | DKFZp686E06161 | Mi-2b | Mi2-BETA | chromodomain helicase DNA binding protein 4 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 9804427 |10220385 |12920132 |
| CTBP1 | BARS | MGC104684 | C-terminal binding protein 1 | - | HPRD,BioGRID | 9650586 |
| DAXX | BING2 | DAP6 | EAP1 | MGC126245 | MGC126246 | death-domain associated protein | - | HPRD,BioGRID | 10669754 |
| DDB2 | DDBB | FLJ34321 | UV-DDB2 | damage-specific DNA binding protein 2, 48kDa | - | HPRD | 8602529 |
| DDX17 | DKFZp761H2016 | P72 | RH70 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | - | HPRD,BioGRID | 15298701 |
| DDX5 | DKFZp686J01190 | G17P1 | HLR1 | HUMP68 | p68 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | Affinity Capture-Western | BioGRID | 15298701 |
| DNMT1 | AIM | CXXC9 | DNMT | FLJ16293 | MCMT | MGC104992 | DNA (cytosine-5-)-methyltransferase 1 | Co-fractionation | BioGRID | 10888886 |
| DNMT1 | AIM | CXXC9 | DNMT | FLJ16293 | MCMT | MGC104992 | DNA (cytosine-5-)-methyltransferase 1 | - | HPRD | 10615135 |
| DNMT3A | DNMT3A2 | M.HsaIIIA | DNA (cytosine-5-)-methyltransferase 3 alpha | - | HPRD,BioGRID | 11350943 |
| DNMT3B | ICF | M.HsaIIIB | DNA (cytosine-5-)-methyltransferase 3 beta | Co-purification | BioGRID | 15148359 |
| DNMT3B | ICF | M.HsaIIIB | DNA (cytosine-5-)-methyltransferase 3 beta | - | HPRD | 12202768 |
| DNMT3B | ICF | M.HsaIIIB | DNA (cytosine-5-)-methyltransferase 3 beta | DNMT3B interacts with HDAC1. | BIND | 15120635 |
| DNMT3L | MGC1090 | DNA (cytosine-5-)-methyltransferase 3-like | - | HPRD,BioGRID | 12177302 |12202768 |
| E2F1 | E2F-1 | RBAP1 | RBBP3 | RBP3 | E2F transcription factor 1 | Affinity Capture-Western | BioGRID | 10734134 |
| E2F4 | E2F-4 | E2F transcription factor 4, p107/p130-binding | - | HPRD,BioGRID | 9724731 |
| EED | HEED | WAIT1 | embryonic ectoderm development | - | HPRD,BioGRID | 10581039 |
| EFCAB6 | DJBP | FLJ23588 | HSCBCIP1 | KIAA1672 | dJ185D5.1 | EF-hand calcium binding domain 6 | Affinity Capture-Western | BioGRID | 12612053 |
| EID2 | CRI2 | EID-2 | MGC20452 | EP300 interacting inhibitor of differentiation 2 | - | HPRD | 12586827 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | ER-alpha interacts with HDAC1. | BIND | 14506733 |
| EVI1 | AML1-EVI-1 | EVI-1 | MDS1-EVI1 | MGC163392 | PRDM3 | ecotropic viral integration site 1 | - | HPRD,BioGRID | 11568182 |
| EZH2 | ENX-1 | EZH1 | KMT6 | MGC9169 | enhancer of zeste homolog 2 (Drosophila) | - | HPRD,BioGRID | 10581039 |
| FKBP3 | FKBP-25 | PPIase | FK506 binding protein 3, 25kDa | - | HPRD,BioGRID | 11532945 |
| FOXG1 | BF1 | BF2 | FHKL3 | FKH2 | FKHL1 | FKHL2 | FKHL3 | FKHL4 | FOXG1A | FOXG1B | FOXG1C | HBF-1 | HBF-2 | HBF-3 | HBF-G2 | HBF2 | HFK1 | HFK2 | HFK3 | KHL2 | QIN | forkhead box G1 | - | HPRD | 11238932 |
| GATA1 | ERYF1 | GF-1 | GF1 | NFE1 | GATA binding protein 1 (globin transcription factor 1) | Affinity Capture-Western | BioGRID | 14668799 |
| GATAD2B | FLJ37346 | KIAA1150 | MGC138257 | MGC138285 | P66beta | RP11-216N14.6 | GATA zinc finger domain containing 2B | Affinity Capture-Western | BioGRID | 11756549 |
| GTF2I | BAP-135 | BAP135 | BTKAP1 | DIWS | FLJ38776 | FLJ56355 | IB291 | SPIN | TFII-I | WBS | WBSCR6 | general transcription factor II, i | Affinity Capture-Western | BioGRID | 12393887 |
| HBP1 | FLJ16340 | HMG-box transcription factor 1 | - | HPRD,BioGRID | 15235594 |
| HCFC1 | CFF | HCF-1 | HCF1 | HFC1 | MGC70925 | VCAF | host cell factor C1 (VP16-accessory protein) | Affinity Capture-MS Affinity Capture-Western | BioGRID | 12670868 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | Affinity Capture-MS | BioGRID | 12920132 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | - | HPRD,BioGRID | 9520398 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | - | HPRD | 10944117 |11302704 |
| HDAC9 | DKFZp779K1053 | HD7 | HDAC | HDAC7 | HDAC7B | HDAC9B | HDAC9FL | HDRP | KIAA0744 | MITR | histone deacetylase 9 | Affinity Capture-Western | BioGRID | 10655483 |
| HDAC9 | DKFZp779K1053 | HD7 | HDAC | HDAC7 | HDAC7B | HDAC9B | HDAC9FL | HDRP | KIAA0744 | MITR | histone deacetylase 9 | An unspecified isoform of HDAC9 (MITR) interacts with HDAC1. This interaction was modelled on a demonstrated interaction between an unspecified isoform of human HDAC9 and HDAC1 from an unspecified species. | BIND | 15711539 |
| HDAC9 | DKFZp779K1053 | HD7 | HDAC | HDAC7 | HDAC7B | HDAC9B | HDAC9FL | HDRP | KIAA0744 | MITR | histone deacetylase 9 | - | HPRD | 12590135 |
| HEY2 | CHF1 | GRIDLOCK | GRL | HERP1 | HESR2 | HRT2 | MGC10720 | bHLHb32 | hairy/enhancer-of-split related with YRPW motif 2 | - | HPRD,BioGRID | 11486045 |
| HIC1 | ZBTB29 | hic-1 | hypermethylated in cancer 1 | - | HPRD | 12052894 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | Reconstituted Complex | BioGRID | 11641274 |
| HIF1AN | DKFZp762F1811 | FIH1 | FLJ20615 | FLJ22027 | hypoxia-inducible factor 1, alpha subunit inhibitor | - | HPRD,BioGRID | 11641274 |
| HMG20B | BRAF25 | BRAF35 | FLJ26127 | HMGX2 | PP7706 | SMARCE1r | SOXL | pp8857 | high-mobility group 20B | Affinity Capture-Western Co-purification | BioGRID | 12032298 |
| HR | ALUNC | AU | HSA277165 | hairless homolog (mouse) | - | HPRD,BioGRID | 11641275 |
| HSPA4 | APG-2 | HS24/P52 | MGC131852 | RY | hsp70 | hsp70RY | heat shock 70kDa protein 4 | Affinity Capture-Western | BioGRID | 11777905 |
| HSPD1 | CPN60 | GROEL | HSP60 | HSP65 | HuCHA60 | SPG13 | heat shock 60kDa protein 1 (chaperonin) | Affinity Capture-Western | BioGRID | 11777905 |
| HUS1 | - | HUS1 checkpoint homolog (S. pombe) | - | HPRD,BioGRID | 10846170 |
| IFRD1 | PC4 | TIS7 | interferon-related developmental regulator 1 | Affinity Capture-Western | BioGRID | 12198164 |
| IKZF1 | Hs.54452 | IK1 | IKAROS | LYF1 | PRO0758 | ZNFN1A1 | hIk-1 | IKAROS family zinc finger 1 (Ikaros) | Affinity Capture-Western | BioGRID | 10357820 |12015313 |
| ING1 | p24ING1c | p33 | p33ING1 | p33ING1b | p47 | p47ING1a | inhibitor of growth family, member 1 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 11784859 |12015309 |
| KAT5 | ESA1 | HTATIP | HTATIP1 | PLIP | TIP | TIP60 | cPLA2 | K(lysine) acetyltransferase 5 | - | HPRD | 11994312 |
| KLF11 | FKLF | FKLF1 | MODY7 | TIEG2 | Tieg3 | Kruppel-like factor 11 | - | HPRD,BioGRID | 11438660 |
| MAD1L1 | HsMAD1 | MAD1 | PIG9 | TP53I9 | TXBP181 | MAD1 mitotic arrest deficient-like 1 (yeast) | Affinity Capture-Western Co-purification | BioGRID | 15388328 |
| MAP3K7IP2 | FLJ21885 | KIAA0733 | TAB2 | mitogen-activated protein kinase kinase kinase 7 interacting protein 2 | Reconstituted Complex | BioGRID | 12150997 |
| MBD2 | DKFZp586O0821 | DMTase | NY-CO-41 | methyl-CpG binding domain protein 2 | - | HPRD,BioGRID | 10471499 |
| MBD3 | - | methyl-CpG binding domain protein 3 | - | HPRD,BioGRID | 12124384 |
| MBD3 | - | methyl-CpG binding domain protein 3 | MBD3 interacts with HDAC1. | BIND | 12124384 |
| MBD3L1 | MBD3L | MGC138263 | MGC138269 | methyl-CpG binding domain protein 3-like 1 | Affinity Capture-Western | BioGRID | 15456747 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | - | HPRD,BioGRID | 12426395 |
| MDS1 | MDS1-EVI1 | PRDM3 | myelodysplasia syndrome 1 | - | HPRD,BioGRID | 11552981 |
| MIER1 | DKFZp781G0451 | ER1 | KIAA1610 | MGC131940 | MGC150640 | MGC150641 | MI-ER1 | RP5-944N15.1 | hMI-ER1 | mesoderm induction early response 1 homolog (Xenopus laevis) | - | HPRD,BioGRID | 12482978 |
| MIZF | DKFZp434F162 | HiNF-P | ZNF743 | MBD2-interacting zinc finger | Affinity Capture-Western | BioGRID | 12665568 |
| MLL | ALL-1 | CXXC7 | FLJ11783 | HRX | HTRX1 | KMT2A | MLL/GAS7 | MLL1A | TET1-MLL | TRX1 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) | Affinity Capture-Western Reconstituted Complex | BioGRID | 12829790 |
| MORF4L2 | KIAA0026 | MORFL2 | MRGX | mortality factor 4 like 2 | - | HPRD | 14506250 |
| MT1G | MGC12386 | MT1 | MT1K | metallothionein 1G | HDAC1 interacts with the MT1G promoter. | BIND | 15735762 |
| MTA1 | - | metastasis associated 1 | - | HPRD,BioGRID | 10840944 |11146623 |
| MTA2 | DKFZp686F2281 | MTA1L1 | PID | metastasis associated 1 family, member 2 | Affinity Capture-MS Affinity Capture-Western Co-purification | BioGRID | 10444591 |11171972 |12374985 |12920132 |
| MXD1 | MAD | MAD1 | MGC104659 | MAX dimerization protein 1 | - | HPRD | 9150133 |9150134 |
| MXD1 | MAD | MAD1 | MGC104659 | MAX dimerization protein 1 | Reconstituted Complex | BioGRID | 11101889 |
| MYOD1 | MYF3 | MYOD | PUM | bHLHc1 | myogenic differentiation 1 | - | HPRD,BioGRID | 11285237 |11684023 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | Affinity Capture-Western Co-purification | BioGRID | 11013263 |11739383 |
| NFIX | NF1A | nuclear factor I/X (CCAAT-binding transcription factor) | Affinity Capture-Western | BioGRID | 11896613 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | - | HPRD,BioGRID | 11931769 |
| NFKBIA | IKBA | MAD-3 | NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | HDAC1 interacts with I-kappa-B-alpha. | BIND | 12972430 |
| NR2F2 | ARP1 | COUP-TFII | COUPTFB | MGC117452 | SVP40 | TFCOUP2 | nuclear receptor subfamily 2, group F, member 2 | - | HPRD,BioGRID | 10704340 |
| NRIP1 | FLJ77253 | RIP140 | nuclear receptor interacting protein 1 | - | HPRD | 11006275|11285237 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | - | HPRD,BioGRID | 11929879 |
| PEX14 | MGC12767 | NAPP2 | Pex14p | dJ734G22.2 | peroxisomal biogenesis factor 14 | - | HPRD | 11909966 |
| PHB | PHB1 | prohibitin | - | HPRD,BioGRID | 12466959 |
| PHF12 | FLJ34122 | KIAA1523 | MGC131914 | PF1 | PHD finger protein 12 | - | HPRD,BioGRID | 11390640 |
| PHF21A | BHC80 | BM-006 | KIAA1696 | PHD finger protein 21A | - | HPRD,BioGRID | 15325272 |
| PIAS3 | FLJ14651 | ZMIZ5 | protein inhibitor of activated STAT, 3 | - | HPRD | 14691252 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | - | HPRD,BioGRID | 11259576 |11430826 |
| PPARD | FAAR | MGC3931 | NR1C2 | NUC1 | NUCI | NUCII | PPAR-beta | PPARB | peroxisome proliferator-activated receptor delta | - | HPRD,BioGRID | 11867749 |
| PTMA | MGC104802 | TMSA | prothymosin, alpha | - | HPRD,BioGRID | 12634383 |
| RAD9A | RAD9 | RAD9 homolog A (S. pombe) | - | HPRD,BioGRID | 10846170 |
| RARB | HAP | NR1B2 | RRB2 | retinoic acid receptor, beta | HDAC1 interacts with RARB (RAR-beta-2) gene promoter sequences. | BIND | 15688037 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | RB interacts with HDAC1. | BIND | 11447271 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | - | HPRD | 9491888 |10888886 |11896613 |11909966 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | The Retinoblastoma protein interacts with Histone Deacetylase 1. | BIND | 10734134 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9491888 |9724731 |10490602 |10615135 |10779361 |11684023 |12466959 |
| RBBP4 | NURF55 | RBAP48 | retinoblastoma binding protein 4 | RbAp48 interacts with HDAC1 | BIND | 10734134 |
| RBBP4 | NURF55 | RBAP48 | retinoblastoma binding protein 4 | - | HPRD | 10734134 |11302704 |12091390 |12943729 |
| RBBP4 | NURF55 | RBAP48 | retinoblastoma binding protein 4 | Affinity Capture-Western Co-purification Reconstituted Complex | BioGRID | 9150133 |9150135 |9520398 |10220385 |10444591 |10471499 |11171972 |11470869 |12091390 |12920132 |12943729 |
| RBBP7 | MGC138867 | MGC138868 | RbAp46 | retinoblastoma binding protein 7 | Affinity Capture-Western Co-purification | BioGRID | 9150135 |10444591 |10471499 |12920132 |
| RBL1 | CP107 | MGC40006 | PRB1 | p107 | retinoblastoma-like 1 (p107) | - | HPRD,BioGRID | 9724731 |
| RBL2 | FLJ26459 | P130 | Rb2 | retinoblastoma-like 2 (p130) | - | HPRD,BioGRID | 10969803 |
| RCOR1 | COREST | KIAA0071 | RCOR | REST corepressor 1 | Affinity Capture-MS Affinity Capture-Western Co-purification | BioGRID | 11171972 |12032298 |
| RECQL4 | RECQ4 | RecQ protein-like 4 | HDAC1 interacts with the RECQL4 (RECQ4) promoter. | BIND | 15674334 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | - | HPRD,BioGRID | 11564889 |
| RFC1 | A1 | MGC51786 | MHCBFB | PO-GA | RECC1 | RFC | RFC140 | replication factor C (activator 1) 1, 145kDa | - | HPRD,BioGRID | 12045192 |
| RFC4 | A1 | MGC27291 | RFC37 | replication factor C (activator 1) 4, 37kDa | - | HPRD | 12045192 |
| RUNX1T1 | AML1T1 | CBFA2T1 | CDR | ETO | MGC2796 | MTG8 | MTG8b | ZMYND2 | runt-related transcription factor 1; translocated to, 1 (cyclin D-related) | Affinity Capture-Western | BioGRID | 11533236 |
| RUNX1T1 | AML1T1 | CBFA2T1 | CDR | ETO | MGC2796 | MTG8 | MTG8b | ZMYND2 | runt-related transcription factor 1; translocated to, 1 (cyclin D-related) | ETO interacts with HDAC1. | BIND | 15333839 |
| RUNX3 | AML2 | CBFA3 | FLJ34510 | MGC16070 | PEBP2aC | runt-related transcription factor 3 | - | HPRD | 15138260 |
| RUVBL2 | CGI-46 | ECP51 | INO80J | REPTIN | RVB2 | TIH2 | TIP48 | TIP49B | RuvB-like 2 (E. coli) | RUVBL2 (reptin) interacts with HDAC1. | BIND | 15829968 |
| SALL1 | HSAL1 | TBS | ZNF794 | sal-like 1 (Drosophila) | - | HPRD | 11836251 |
| SAP130 | FLJ12761 | Sin3A-associated protein, 130kDa | Affinity Capture-Western | BioGRID | 12724404 |
| SAP18 | 2HOR0202 | MGC27131 | SAP18p | Sin3A-associated protein, 18kDa | - | HPRD | 9150135 |
| SAP30 | - | Sin3A-associated protein, 30kDa | - | HPRD,BioGRID | 9651585 |
| SATB1 | - | SATB homeobox 1 | - | HPRD,BioGRID | 12374985 |
| SETDB1 | ESET | KG1T | KIAA0067 | KMT1E | SET domain, bifurcated 1 | - | HPRD | 12398767 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | The transcriptional repressor mSin3A interacts with the histone deacetylase 1 protein. | BIND | 9150133 |9150134 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | Affinity Capture-MS Affinity Capture-Western Co-fractionation Co-purification | BioGRID | 9520398 |9804427 |10220385 |10444591 |10640275 |11171972 |11784859 |11931768 |12091390 |12374985 |12398767 |12724404 |12920132 |12943729 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | - | HPRD | 11013263 |
| SIN3B | KIAA0700 | SIN3 homolog B, transcription regulator (yeast) | - | HPRD | 11959842 |
| SIN3B | KIAA0700 | SIN3 homolog B, transcription regulator (yeast) | Affinity Capture-Western Co-purification | BioGRID | 10357820 |10444591 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | - | HPRD,BioGRID | 10199400 |
| SMARCA2 | BAF190 | BRM | FLJ36757 | MGC74511 | SNF2 | SNF2L2 | SNF2LA | SWI2 | Sth1p | hBRM | hSNF2a | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | Co-purification | BioGRID | 11238380 |
| SMARCB1 | BAF47 | INI1 | RDT | SNF5 | SNF5L1 | Sfh1p | Snr1 | hSNFS | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 | Co-purification | BioGRID | 11238380 |
| SP1 | - | Sp1 transcription factor | Affinity Capture-Western | BioGRID | 11896613 |12091390 |12176973 |12847090 |
| SP1 | - | Sp1 transcription factor | - | HPRD | 11896613 |11909966 |
| SP3 | DKFZp686O1631 | SPR-2 | Sp3 transcription factor | - | HPRD,BioGRID | 12176973 |
| SPEN | KIAA0929 | MINT | RP1-134O19.1 | SHARP | spen homolog, transcriptional regulator (Drosophila) | - | HPRD,BioGRID | 11331609 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | Stat3 interacts with HDAC1. | BIND | 15653507 |
| SUDS3 | FLJ00052 | MGC104711 | SAP45 | SDS3 | suppressor of defective silencing 3 homolog (S. cerevisiae) | Affinity Capture-Western | BioGRID | 11909966 |12724404 |
| SUV39H1 | KMT1A | MG44 | SUV39H | suppressor of variegation 3-9 homolog 1 (Drosophila) | - | HPRD,BioGRID | 11788710 |
| TAL1 | SCL | TCL5 | bHLHa17 | tal-1 | T-cell acute lymphocytic leukemia 1 | - | HPRD | 10688671 |
| TAL1 | SCL | TCL5 | bHLHa17 | tal-1 | T-cell acute lymphocytic leukemia 1 | Affinity Capture-MS | BioGRID | 16407974 |
| TGIF1 | HPE4 | MGC39747 | MGC5066 | TGIF | TGFB-induced factor homeobox 1 | Affinity Capture-Western | BioGRID | 10995736 |11427533 |
| TGIF2 | - | TGFB-induced factor homeobox 2 | - | HPRD,BioGRID | 11427533 |
| TLE4 | BCE-1 | BCE1 | E(spI) | ESG | ESG4 | GRG4 | transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) | Affinity Capture-Western | BioGRID | 11266540 |
| TNFSF10 | APO2L | Apo-2L | CD253 | TL2 | TRAIL | tumor necrosis factor (ligand) superfamily, member 10 | HDAC1 interacts with the TNFSF10 promoter. | BIND | 15619633 |
| TOP2A | TOP2 | TP2A | topoisomerase (DNA) II alpha 170kDa | - | HPRD,BioGRID | 11136718 |
| TOP2B | TOPIIB | top2beta | topoisomerase (DNA) II beta 180kDa | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 11062478 |11136718 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Affinity Capture-Western | BioGRID | 12426395 |
| TPD52L1 | D53 | MGC8556 | TPD52L2 | hD53 | tumor protein D52-like 1 | Two-hybrid | BioGRID | 16169070 |
| TRIM27 | RFP | RNF76 | tripartite motif-containing 27 | - | HPRD | 14530259 |
| TXNIP | EST01027 | HHCPA78 | THIF | VDUP1 | thioredoxin interacting protein | - | HPRD,BioGRID | 12821938 |
| VHL | HRCA1 | RCA1 | VHL1 | von Hippel-Lindau tumor suppressor | Reconstituted Complex | BioGRID | 11641274 |
| YY1 | DELTA | INO80S | NF-E1 | UCRBP | YIN-YANG-1 | YY1 transcription factor | Reconstituted Complex | BioGRID | 9346952 |
| ZBTB16 | PLZF | ZNF145 | zinc finger and BTB domain containing 16 | - | HPRD | 9627120 |15467736 |
| ZBTB16 | PLZF | ZNF145 | zinc finger and BTB domain containing 16 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9627120 |9765306 |15467736 |
| ZBTB16 | PLZF | ZNF145 | zinc finger and BTB domain containing 16 | HDAC1 interacts with PLZF. This interaction was modelled on a demonstrated interaction between HDAC1 and PLZF from unspecified species. | BIND | 9486654 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG NOTCH SIGNALING PATHWAY | 47 | 35 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG CHRONIC MYELOID LEUKEMIA | 73 | 59 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CARM ER PATHWAY | 35 | 27 | All SZGR 2.0 genes in this pathway |
| BIOCARTA G1 PATHWAY | 28 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA VDR PATHWAY | 12 | 7 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MTA3 PATHWAY | 19 | 10 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PITX2 PATHWAY | 15 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MEF2D PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA WNT PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID NOTCH PATHWAY | 59 | 49 | All SZGR 2.0 genes in this pathway |
| PID E2F PATHWAY | 74 | 48 | All SZGR 2.0 genes in this pathway |
| PID PS1 PATHWAY | 46 | 39 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSI PATHWAY | 66 | 50 | All SZGR 2.0 genes in this pathway |
| PID TELOMERASE PATHWAY | 68 | 48 | All SZGR 2.0 genes in this pathway |
| PID REG GR PATHWAY | 82 | 60 | All SZGR 2.0 genes in this pathway |
| PID RANBP2 PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| PID AR TF PATHWAY | 53 | 38 | All SZGR 2.0 genes in this pathway |
| PID IL3 PATHWAY | 27 | 19 | All SZGR 2.0 genes in this pathway |
| PID ERA GENOMIC PATHWAY | 65 | 37 | All SZGR 2.0 genes in this pathway |
| PID RETINOIC ACID PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| PID HEDGEHOG GLI PATHWAY | 48 | 35 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN NUC PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID MYC REPRESS PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| PID RB 1PATHWAY | 65 | 46 | All SZGR 2.0 genes in this pathway |
| PID HES HEY PATHWAY | 48 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME G0 AND EARLY G1 | 25 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | 46 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH1 | 70 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | 38 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | 81 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | 132 | 101 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH | 103 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | 63 | 42 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 UP | 201 | 125 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP UP | 265 | 158 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR DN | 64 | 39 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR DN | 75 | 50 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 8 | 36 | 28 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER ACOX1 UP | 64 | 40 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA | 176 | 122 | All SZGR 2.0 genes in this pathway |
| XU RESPONSE TO TRETINOIN AND NSC682994 UP | 17 | 13 | All SZGR 2.0 genes in this pathway |
| NGUYEN NOTCH1 TARGETS UP | 29 | 23 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY TBH AND H2O2 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX DN | 80 | 49 | All SZGR 2.0 genes in this pathway |
| RHODES CANCER META SIGNATURE | 64 | 47 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C1 | 72 | 45 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR UP | 294 | 199 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| GU PDEF TARGETS UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| SETLUR PROSTATE CANCER TMPRSS2 ERG FUSION UP | 67 | 48 | All SZGR 2.0 genes in this pathway |
| KESHELAVA MULTIPLE DRUG RESISTANCE | 88 | 56 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS UP | 295 | 155 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-410 | 90 | 96 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.