Gene Page: HLA-B
Summary ?
| GeneID | 3106 |
| Symbol | HLA-B |
| Synonyms | AS|Bw-47|Bw-50|HLAB|SPDA1 |
| Description | major histocompatibility complex, class I, B |
| Reference | MIM:142830|HGNC:HGNC:4932|Ensembl:ENSG00000234745|Ensembl:ENST00000359635|Ensembl:ENST00000435618|Ensembl:ENST00000450871|HPRD:00828|Vega:OTTHUMG00000031153 |
| Gene type | protein-coding |
| Map location | 6p21.3 |
| Sherlock p-value | 0.381 |
| Fetal beta | -0.622 |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Frontal Cortex BA9 Hippocampus Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.5489 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4081552 | chr6 | 31353688 | HLA-B | 3106 | 0.16 | cis | ||
| rs10495652 | chr2 | 17139288 | HLA-B | 3106 | 0.04 | trans | ||
| rs6531271 | chr2 | 17139932 | HLA-B | 3106 | 0.04 | trans | ||
| rs12620194 | chr2 | 17148003 | HLA-B | 3106 | 0.04 | trans | ||
| rs17503903 | chr2 | 51039461 | HLA-B | 3106 | 0.1 | trans | ||
| rs890527 | chr3 | 140774852 | HLA-B | 3106 | 0.04 | trans | ||
| rs1678893 | chr5 | 31763244 | HLA-B | 3106 | 0.14 | trans | ||
| rs7741955 | chr6 | 84829621 | HLA-B | 3106 | 0.15 | trans | ||
| rs12524136 | chr6 | 84935440 | HLA-B | 3106 | 0.05 | trans | ||
| rs9397692 | chr6 | 154487314 | HLA-B | 3106 | 0.2 | trans | ||
| rs10509151 | chr10 | 63358651 | HLA-B | 3106 | 0.15 | trans | ||
| rs6488258 | chr12 | 10252207 | HLA-B | 3106 | 0.17 | trans | ||
| rs4769240 | chr13 | 23697920 | HLA-B | 3106 | 0.19 | trans | ||
| rs16979927 | chr20 | 55019249 | HLA-B | 3106 | 0 | trans | ||
| rs2832206 | chr21 | 30504653 | HLA-B | 3106 | 0.15 | trans | ||
| rs17347454 | chrX | 23347700 | HLA-B | 3106 | 0.01 | trans | ||
| rs689365 | chrX | 97074769 | HLA-B | 3106 | 0.15 | trans |
Section II. Transcriptome annotation
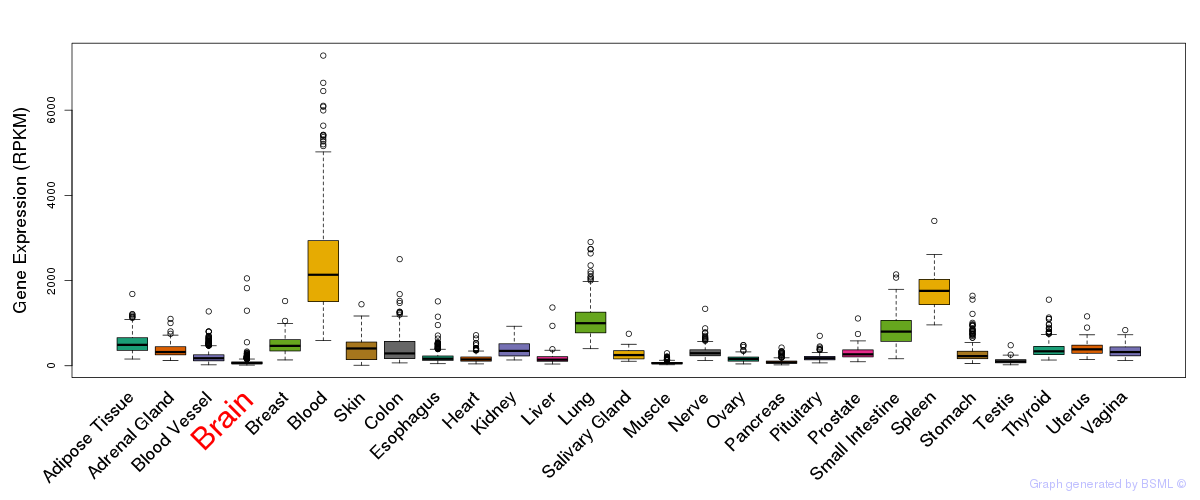
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003674 | molecular_function | ND | - | |
| GO:0005515 | protein binding | IPI | 17353931 | |
| GO:0032393 | MHC class I receptor activity | NAS | 518865 |2715640 |8420828 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | IEA | - | |
| GO:0019882 | antigen processing and presentation | IEA | - | |
| GO:0008150 | biological_process | ND | 10203017 | |
| GO:0006955 | immune response | IEA | - | |
| GO:0006955 | immune response | NAS | 518865 |2715640 |8420828 | |
| GO:0006952 | defense response | TAS | 3011411 | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005575 | cellular_component | ND | - | |
| GO:0005624 | membrane fraction | TAS | 3011411 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | EXP | 15304001 | |
| GO:0005887 | integral to plasma membrane | NAS | 518865 |2715640 |8420828 | |
| GO:0042612 | MHC class I protein complex | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AHCY | SAHH | S-adenosylhomocysteine hydrolase | Affinity Capture-MS | BioGRID | 17353931 |
| B2M | - | beta-2-microglobulin | - | HPRD | 2450918 |
| CD8A | CD8 | Leu2 | MAL | p32 | CD8a molecule | - | HPRD,BioGRID | 9331948 |10809759|10809759 |
| EZR | CVIL | CVL | DKFZp762H157 | FLJ26216 | MGC1584 | VIL2 | ezrin | Affinity Capture-MS | BioGRID | 17353931 |
| KIR3DL1 | CD158E1 | KIR | MGC119726 | MGC119728 | MGC126589 | MGC126591 | NKAT3 | NKB1 | NKB1B | killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 1 | in vivo | BioGRID | 8046332 |
| KIR3DL1 | CD158E1 | KIR | MGC119726 | MGC119728 | MGC126589 | MGC126591 | NKAT3 | NKB1 | NKB1B | killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 1 | - | HPRD | 12134147 |
| KIR3DS1 | CD158E2 | KIR-123FM | KIR-G1 | MGC119726 | MGC119728 | MGC125316 | NKAT10 | killer cell immunoglobulin-like receptor, three domains, short cytoplasmic tail, 1 | Phenotypic Enhancement | BioGRID | 12134147 |
| KLRD1 | CD94 | killer cell lectin-like receptor subfamily D, member 1 | - | HPRD | 8046333 |
| LILRB1 | CD85 | CD85J | FLJ37515 | ILT2 | LIR-1 | LIR1 | MIR-7 | MIR7 | leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 | - | HPRD,BioGRID | 12853576 |
| LILRB2 | CD85D | ILT4 | LILRA6 | LIR-2 | LIR2 | MIR-10 | MIR10 | leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2 | - | HPRD,BioGRID | 9531263 |12853576 |
| LILRB2 | CD85D | ILT4 | LILRA6 | LIR-2 | LIR2 | MIR-10 | MIR10 | leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2 | - | HPRD | 12853576 |
| MAGEA1 | MAGE1 | MGC9326 | melanoma antigen family A, 1 (directs expression of antigen MZ2-E) | - | HPRD,BioGRID | 10363990 |
| MSN | - | moesin | Affinity Capture-MS | BioGRID | 17353931 |
| PAICS | ADE2 | ADE2H1 | AIRC | DKFZp781N1372 | MGC1343 | MGC5024 | PAIS | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase | Affinity Capture-MS | BioGRID | 17353931 |
| PFAS | FGAMS | FGARAT | KIAA0361 | PURL | phosphoribosylformylglycinamidine synthase | Affinity Capture-MS | BioGRID | 17353931 |
| PLS3 | T-plastin | plastin 3 (T isoform) | Affinity Capture-MS | BioGRID | 17353931 |
| PSMD1 | MGC133040 | MGC133041 | P112 | Rpn2 | S1 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 | Affinity Capture-MS | BioGRID | 17353931 |
| RUVBL1 | ECP54 | INO80H | NMP238 | PONTIN | Pontin52 | RVB1 | TIH1 | TIP49 | TIP49A | RuvB-like 1 (E. coli) | Affinity Capture-MS | BioGRID | 17353931 |
| TARS | MGC9344 | ThrRS | threonyl-tRNA synthetase | Affinity Capture-MS | BioGRID | 17353931 |
| TRA@ | FLJ22602 | MGC117436 | MGC22624 | MGC23964 | MGC71411 | TCRA | TCRD | TRA | T cell receptor alpha locus | - | HPRD,BioGRID | 12530975 |
| TRIM28 | FLJ29029 | KAP1 | RNF96 | TF1B | TIF1B | tripartite motif-containing 28 | Affinity Capture-MS | BioGRID | 17353931 |
| VARS | G7A | VARS1 | VARS2 | valyl-tRNA synthetase | Affinity Capture-MS | BioGRID | 17353931 |
| VCP | IBMPFD | MGC131997 | MGC148092 | MGC8560 | TERA | p97 | valosin-containing protein | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| KEGG ANTIGEN PROCESSING AND PRESENTATION | 89 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG NATURAL KILLER CELL MEDIATED CYTOTOXICITY | 137 | 92 | All SZGR 2.0 genes in this pathway |
| KEGG TYPE I DIABETES MELLITUS | 44 | 38 | All SZGR 2.0 genes in this pathway |
| KEGG AUTOIMMUNE THYROID DISEASE | 53 | 49 | All SZGR 2.0 genes in this pathway |
| KEGG ALLOGRAFT REJECTION | 38 | 34 | All SZGR 2.0 genes in this pathway |
| KEGG GRAFT VERSUS HOST DISEASE | 42 | 31 | All SZGR 2.0 genes in this pathway |
| KEGG VIRAL MYOCARDITIS | 73 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | 76 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME ENDOSOMAL VACUOLAR PATHWAY | 9 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME ER PHAGOSOME PATHWAY | 61 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | 70 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON GAMMA SIGNALING | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON ALPHA BETA SIGNALING | 64 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING | 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA DN | 116 | 79 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL DN | 118 | 79 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| WIELAND UP BY HBV INFECTION | 101 | 66 | All SZGR 2.0 genes in this pathway |
| SANA RESPONSE TO IFNG UP | 78 | 50 | All SZGR 2.0 genes in this pathway |
| SANA TNF SIGNALING UP | 83 | 56 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP | 207 | 145 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| GALE APL WITH FLT3 MUTATED DN | 17 | 10 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| HU GENOTOXIN ACTION DIRECT VS INDIRECT 24HR | 55 | 38 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| MARCHINI TRABECTEDIN RESISTANCE DN | 49 | 34 | All SZGR 2.0 genes in this pathway |
| KIM LRRC3B TARGETS | 30 | 24 | All SZGR 2.0 genes in this pathway |
| PELLICCIOTTA HDAC IN ANTIGEN PRESENTATION DN | 49 | 38 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| CAMPS COLON CANCER COPY NUMBER DN | 74 | 34 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR UP | 174 | 96 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE UP | 126 | 92 | All SZGR 2.0 genes in this pathway |
| GAURNIER PSMD4 TARGETS | 73 | 55 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN | 216 | 143 | All SZGR 2.0 genes in this pathway |
| HUPER BREAST BASAL VS LUMINAL DN | 59 | 44 | All SZGR 2.0 genes in this pathway |
| WUNDER INFLAMMATORY RESPONSE AND CHOLESTEROL UP | 58 | 38 | All SZGR 2.0 genes in this pathway |
| HOFFMANN SMALL PRE BII TO IMMATURE B LYMPHOCYTE DN | 50 | 33 | All SZGR 2.0 genes in this pathway |
| LEE EARLY T LYMPHOCYTE DN | 57 | 36 | All SZGR 2.0 genes in this pathway |
| KUROKAWA LIVER CANCER EARLY RECURRENCE DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| STAMBOLSKY TARGETS OF MUTATED TP53 DN | 50 | 24 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 UP | 140 | 94 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |