Gene Page: HNRNPU
Summary ?
| GeneID | 3192 |
| Symbol | HNRNPU |
| Synonyms | HNRNPU-AS1|HNRPU|SAF-A|SAFA|U21.1|hnRNP U |
| Description | heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| Reference | MIM:602869|HGNC:HGNC:5048|Ensembl:ENSG00000153187|Ensembl:ENSG00000188206|HPRD:04185|Vega:OTTHUMG00000040396 |
| Gene type | protein-coding |
| Map location | 1q44 |
| Sherlock p-value | 0.057 |
| Fetal beta | 1.485 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24517768 | 1 | 245028012 | HNRNPU | 9.5E-11 | -0.01 | 4.49E-7 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10035168 | chr5 | 5974295 | HNRNPU | 3192 | 0.16 | trans |
Section II. Transcriptome annotation
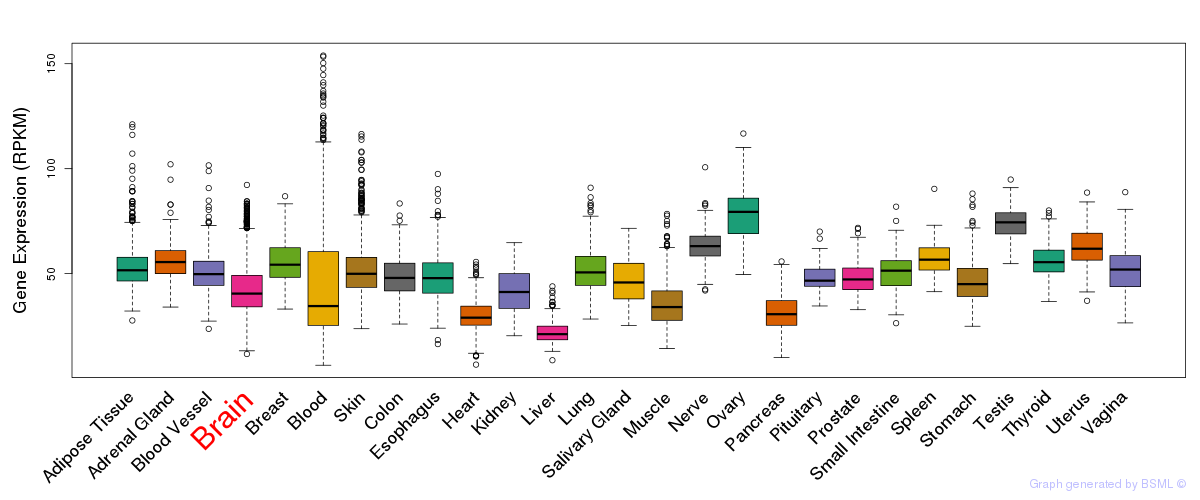
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MAB21L2 | 0.95 | 0.07 |
| IRX2 | 0.84 | -0.11 |
| IRX1 | 0.67 | -0.06 |
| WFIKKN2 | 0.58 | 0.08 |
| NRP1 | 0.54 | 0.04 |
| ROBO3 | 0.54 | 0.13 |
| PRDM6 | 0.49 | 0.06 |
| TMC5 | 0.42 | 0.08 |
| ROR2 | 0.38 | 0.18 |
| MXRA5 | 0.36 | 0.15 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| WDR53 | -0.11 | -0.21 |
| AP1S2 | -0.10 | -0.20 |
| RALYL | -0.10 | -0.21 |
| ZNF860 | -0.10 | -0.32 |
| RPL36AL | -0.09 | -0.21 |
| ZNF230 | -0.09 | -0.29 |
| IER5L | -0.09 | -0.10 |
| GIN1 | -0.09 | -0.20 |
| DNAJB6 | -0.09 | -0.29 |
| PDCL3 | -0.09 | -0.26 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003676 | nucleic acid binding | IEA | - | |
| GO:0003677 | DNA binding | TAS | 1628625 | |
| GO:0003723 | RNA binding | TAS | 1628625 | |
| GO:0005515 | protein binding | IPI | 17220478 | |
| GO:0005524 | ATP binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000398 | nuclear mRNA splicing, via spliceosome | EXP | 12226669 | |
| GO:0008380 | RNA splicing | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005681 | spliceosome | IEA | - | |
| GO:0009986 | cell surface | IEA | - | |
| GO:0030529 | ribonucleoprotein complex | IEA | - | |
| GO:0030530 | heterogeneous nuclear ribonucleoprotein complex | TAS | 7509195 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTB | PS1TP5BP1 | actin, beta | ACTB (beta-actin) interacts with HNRPU (hnRNP U). | BIND | 15711563 |
| CDK7 | CAK1 | CDKN7 | MO15 | STK1 | p39MO15 | cyclin-dependent kinase 7 | Affinity Capture-Western | BioGRID | 10490622 |
| CR2 | C3DR | CD21 | SLEB9 | complement component (3d/Epstein Barr virus) receptor 2 | p120RNP interacts with CR2. | BIND | 7753047 |
| ELL | C19orf17 | DKFZp434I1916 | ELL1 | Men | elongation factor RNA polymerase II | - | HPRD | 10490622 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | - | HPRD,BioGRID | 11909954 |
| GRIN1 | NMDA1 | NMDAR1 | NR1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | - | HPRD | 10862698 |
| GTF2F1 | BTF4 | RAP74 | TF2F1 | TFIIF | general transcription factor IIF, polypeptide 1, 74kDa | Affinity Capture-Western | BioGRID | 10490622 |
| GTF2H1 | BTF2 | TFB1 | TFIIH | general transcription factor IIH, polypeptide 1, 62kDa | - | HPRD | 10490622 |
| NDN | HsT16328 | PWCR | necdin homolog (mouse) | - | HPRD,BioGRID | 11813259 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | - | HPRD,BioGRID | 9353307 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | Affinity Capture-Western | BioGRID | 10490622 |
| POU3F4 | BRAIN-4 | BRN4 | DFN3 | DFNX2 | OTF9 | POU class 3 homeobox 4 | - | HPRD,BioGRID | 9105675 |
| PRMT1 | ANM1 | HCP1 | HRMT1L2 | IR1B4 | protein arginine methyltransferase 1 | - | HPRD,BioGRID | 11850402 |
| SFN | YWHAS | stratifin | Affinity Capture-MS | BioGRID | 15778465 |
| SFRS12 | DKFZp564B176 | MGC133045 | SRrp508 | SRrp86 | splicing factor, arginine/serine-rich 12 | - | HPRD | 14559993 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SPLICEOSOME | 128 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | 140 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING | 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING | 111 | 58 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS UP | 214 | 134 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| OUELLET OVARIAN CANCER INVASIVE VS LMP UP | 117 | 85 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD UP | 64 | 39 | All SZGR 2.0 genes in this pathway |
| DACOSTA LOW DOSE UV RESPONSE VIA ERCC3 XPCS UP | 18 | 10 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP | 309 | 199 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| STARK HYPPOCAMPUS 22Q11 DELETION UP | 53 | 40 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 DN | 169 | 102 | All SZGR 2.0 genes in this pathway |
| DORSAM HOXA9 TARGETS UP | 35 | 25 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| BROCKE APOPTOSIS REVERSED BY IL6 | 144 | 98 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH REARRANGEMENTS DN | 48 | 26 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C2 | 54 | 39 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D6 | 37 | 25 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| ZHOU TNF SIGNALING 30MIN | 54 | 36 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS UP | 66 | 43 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER POOR SURVIVAL DN | 22 | 15 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR DN | 185 | 116 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER UP | 83 | 63 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S2 | 115 | 74 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| STAMBOLSKY TARGETS OF MUTATED TP53 UP | 49 | 35 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 DN | 92 | 60 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN VINCRISTINE RESISTANCE B ALL DN | 15 | 10 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN VINCRISTINE RESISTANCE ALL DN | 19 | 11 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 86 | 93 | 1A,m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-101 | 855 | 861 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-122 | 475 | 482 | 1A,m8 | hsa-miR-122a | UGGAGUGUGACAAUGGUGUUUGU |
| miR-132/212 | 55 | 61 | m8 | hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC |
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| miR-135 | 1421 | 1427 | m8 | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-142-5p | 1102 | 1108 | 1A | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-144 | 854 | 861 | 1A,m8 | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-150 | 1307 | 1313 | 1A | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| miR-186 | 62 | 68 | 1A | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU | ||||
| miR-19 | 1525 | 1532 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-200bc/429 | 234 | 240 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-21 | 901 | 907 | 1A | hsa-miR-21brain | UAGCUUAUCAGACUGAUGUUGA |
| hsa-miR-590 | GAGCUUAUUCAUAAAAGUGCAG | ||||
| miR-214 | 979 | 986 | 1A,m8 | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-224 | 269 | 275 | m8 | hsa-miR-224 | CAAGUCACUAGUGGUUCCGUUUA |
| miR-23 | 266 | 272 | m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-25/32/92/363/367 | 1040 | 1046 | 1A | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-30-3p | 741 | 747 | 1A | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-323 | 266 | 272 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-330 | 309 | 315 | m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-409-3p | 85 | 91 | m8 | hsa-miR-409-3p | CGAAUGUUGCUCGGUGAACCCCU |
| miR-433-3p | 439 | 445 | 1A | hsa-miR-433brain | AUCAUGAUGGGCUCCUCGGUGU |
| miR-495 | 1033 | 1039 | m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-496 | 655 | 661 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-539 | 28 | 34 | m8 | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
| hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU | ||||
| miR-542-3p | 383 | 389 | 1A | hsa-miR-542-3p | UGUGACAGAUUGAUAACUGAAA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.