Gene Page: CXCL10
Summary ?
| GeneID | 3627 |
| Symbol | CXCL10 |
| Synonyms | C7|IFI10|INP10|IP-10|SCYB10|crg-2|gIP-10|mob-1 |
| Description | C-X-C motif chemokine ligand 10 |
| Reference | MIM:147310|HGNC:HGNC:10637|Ensembl:ENSG00000169245|HPRD:00930|Vega:OTTHUMG00000160887 |
| Gene type | protein-coding |
| Map location | 4q21 |
| Pascal p-value | 0.067 |
| Fetal beta | -0.622 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10915489 | chr1 | 4321466 | CXCL10 | 3627 | 0.09 | trans | ||
| rs12036740 | chr1 | 58432174 | CXCL10 | 3627 | 0.04 | trans | ||
| rs10789056 | chr1 | 58457827 | CXCL10 | 3627 | 0.18 | trans | ||
| rs12041093 | chr1 | 58462509 | CXCL10 | 3627 | 0.01 | trans | ||
| rs12023109 | chr1 | 58482942 | CXCL10 | 3627 | 0.01 | trans | ||
| rs473019 | chr1 | 72296394 | CXCL10 | 3627 | 3.789E-4 | trans | ||
| rs17359517 | chr1 | 115381897 | CXCL10 | 3627 | 0.11 | trans | ||
| rs1342707 | chr1 | 147264296 | CXCL10 | 3627 | 9.186E-9 | trans | ||
| rs12076447 | chr1 | 159552549 | CXCL10 | 3627 | 5.311E-5 | trans | ||
| rs10752920 | chr1 | 183783202 | CXCL10 | 3627 | 0.13 | trans | ||
| rs10494571 | chr1 | 183930700 | CXCL10 | 3627 | 9.805E-6 | trans | ||
| rs6656512 | chr1 | 223644438 | CXCL10 | 3627 | 0.08 | trans | ||
| rs4027083 | chr1 | 234452908 | CXCL10 | 3627 | 0.05 | trans | ||
| rs2907143 | chr1 | 243147434 | CXCL10 | 3627 | 0.05 | trans | ||
| rs10495652 | chr2 | 17139288 | CXCL10 | 3627 | 0.09 | trans | ||
| rs6531271 | chr2 | 17139932 | CXCL10 | 3627 | 0.09 | trans | ||
| rs12620194 | chr2 | 17148003 | CXCL10 | 3627 | 0.09 | trans | ||
| rs16987384 | chr2 | 20328893 | CXCL10 | 3627 | 0.09 | trans | ||
| rs17042744 | chr2 | 21824821 | CXCL10 | 3627 | 0 | trans | ||
| rs918897 | chr2 | 21825859 | CXCL10 | 3627 | 0.08 | trans | ||
| rs2113625 | chr2 | 21828617 | CXCL10 | 3627 | 0 | trans | ||
| rs17680142 | chr2 | 21849670 | CXCL10 | 3627 | 0 | trans | ||
| rs6547745 | chr2 | 21852551 | CXCL10 | 3627 | 0 | trans | ||
| rs6758775 | chr2 | 21887142 | CXCL10 | 3627 | 0.02 | trans | ||
| rs17042868 | chr2 | 21895688 | CXCL10 | 3627 | 0 | trans | ||
| rs11893111 | chr2 | 21910980 | CXCL10 | 3627 | 0 | trans | ||
| rs4488633 | chr2 | 35142820 | CXCL10 | 3627 | 0.16 | trans | ||
| rs6760426 | chr2 | 66175535 | CXCL10 | 3627 | 0.15 | trans | ||
| rs17546660 | chr2 | 66218021 | CXCL10 | 3627 | 0.12 | trans | ||
| rs2218030 | chr2 | 184261431 | CXCL10 | 3627 | 0.07 | trans | ||
| rs16824418 | chr2 | 184275364 | CXCL10 | 3627 | 0.19 | trans | ||
| rs17033273 | chr3 | 10676696 | CXCL10 | 3627 | 0.01 | trans | ||
| rs277655 | chr3 | 100044586 | CXCL10 | 3627 | 0.14 | trans | ||
| rs16847252 | chr3 | 163481290 | CXCL10 | 3627 | 2.588E-5 | trans | ||
| rs2215847 | chr4 | 25067686 | CXCL10 | 3627 | 0.12 | trans | ||
| rs591585 | chr4 | 43424979 | CXCL10 | 3627 | 0.07 | trans | ||
| rs6551878 | chr4 | 65690588 | CXCL10 | 3627 | 0.11 | trans | ||
| rs17622486 | chr4 | 115785097 | CXCL10 | 3627 | 0.05 | trans | ||
| rs298465 | chr4 | 139260859 | CXCL10 | 3627 | 0.19 | trans | ||
| rs11953760 | chr5 | 9842943 | CXCL10 | 3627 | 0.09 | trans | ||
| rs7380779 | chr5 | 34436068 | CXCL10 | 3627 | 0 | trans | ||
| rs7380792 | chr5 | 34436187 | CXCL10 | 3627 | 0 | trans | ||
| rs10074060 | chr5 | 34437321 | CXCL10 | 3627 | 0.05 | trans | ||
| rs7442700 | chr5 | 34493693 | CXCL10 | 3627 | 0.06 | trans | ||
| rs4516856 | chr5 | 43889206 | CXCL10 | 3627 | 0.05 | trans | ||
| rs31304 | chr5 | 52942082 | CXCL10 | 3627 | 0.01 | trans | ||
| rs10514893 | chr5 | 59657694 | CXCL10 | 3627 | 0.11 | trans | ||
| rs10514894 | chr5 | 59658797 | CXCL10 | 3627 | 0.18 | trans | ||
| rs10515260 | chr5 | 97076547 | CXCL10 | 3627 | 2.687E-12 | trans | ||
| rs10515267 | chr5 | 100128201 | CXCL10 | 3627 | 0.03 | trans | ||
| rs17780121 | chr5 | 100168511 | CXCL10 | 3627 | 0.05 | trans | ||
| rs17780992 | chr5 | 100195770 | CXCL10 | 3627 | 0.05 | trans | ||
| rs9313842 | chr5 | 159690030 | CXCL10 | 3627 | 0.05 | trans | ||
| rs6920921 | chr6 | 8884160 | CXCL10 | 3627 | 0.11 | trans | ||
| rs2327748 | chr6 | 14082533 | CXCL10 | 3627 | 0.01 | trans | ||
| rs12195984 | 0 | CXCL10 | 3627 | 0.14 | trans | |||
| rs10498827 | chr6 | 65470694 | CXCL10 | 3627 | 0.08 | trans | ||
| rs16869716 | chr6 | 71743471 | CXCL10 | 3627 | 9.782E-8 | trans | ||
| rs2093506 | chr6 | 154232014 | CXCL10 | 3627 | 0.06 | trans | ||
| rs6455821 | chr6 | 162825101 | CXCL10 | 3627 | 0.06 | trans | ||
| rs6956395 | chr7 | 28072244 | CXCL10 | 3627 | 0.01 | trans | ||
| rs6462122 | chr7 | 29137630 | CXCL10 | 3627 | 0.01 | trans | ||
| rs7776597 | chr7 | 51015581 | CXCL10 | 3627 | 0.08 | trans | ||
| rs3937454 | chr7 | 152676939 | CXCL10 | 3627 | 0.19 | trans | ||
| rs7790094 | chr7 | 152786528 | CXCL10 | 3627 | 0.02 | trans | ||
| rs4410820 | chr7 | 152853659 | CXCL10 | 3627 | 0.05 | trans | ||
| rs17075051 | chr8 | 5761144 | CXCL10 | 3627 | 0.02 | trans | ||
| rs17075079 | chr8 | 5765448 | CXCL10 | 3627 | 0.05 | trans | ||
| rs10503341 | chr8 | 5775097 | CXCL10 | 3627 | 0.18 | trans | ||
| rs9325708 | chr8 | 12402940 | CXCL10 | 3627 | 7.414E-4 | trans | ||
| rs7817084 | chr8 | 43242956 | CXCL10 | 3627 | 0.14 | trans | ||
| rs10959381 | chr9 | 10936134 | CXCL10 | 3627 | 0.11 | trans | ||
| rs12344698 | chr9 | 16750142 | CXCL10 | 3627 | 0.07 | trans | ||
| rs17376956 | chr10 | 11179948 | CXCL10 | 3627 | 0.17 | trans | ||
| rs3802522 | chr10 | 28341865 | CXCL10 | 3627 | 4.127E-7 | trans | ||
| rs10508730 | chr10 | 28403391 | CXCL10 | 3627 | 4.127E-7 | trans | ||
| rs16936983 | chr10 | 37871050 | CXCL10 | 3627 | 0.14 | trans | ||
| rs1344342 | chr10 | 37958911 | CXCL10 | 3627 | 0.14 | trans | ||
| rs1108733 | chr10 | 37993286 | CXCL10 | 3627 | 0.14 | trans | ||
| rs4917410 | chr10 | 95872539 | CXCL10 | 3627 | 0.17 | trans | ||
| rs7089703 | chr10 | 119182182 | CXCL10 | 3627 | 0.06 | trans | ||
| rs11246002 | chr11 | 211446 | CXCL10 | 3627 | 0.16 | trans | ||
| rs10743139 | chr11 | 10425326 | CXCL10 | 3627 | 0.02 | trans | ||
| rs2508654 | chr11 | 120405519 | CXCL10 | 3627 | 0.02 | trans | ||
| rs1944608 | chr11 | 123091803 | CXCL10 | 3627 | 0.09 | trans | ||
| rs17127157 | chr11 | 123092323 | CXCL10 | 3627 | 0.09 | trans | ||
| rs10893162 | chr11 | 124076268 | CXCL10 | 3627 | 0.09 | trans | ||
| rs11219545 | chr11 | 124136438 | CXCL10 | 3627 | 0.09 | trans | ||
| rs11219646 | chr11 | 124279239 | CXCL10 | 3627 | 0.04 | trans | ||
| rs7102988 | chr11 | 130134440 | CXCL10 | 3627 | 0.16 | trans | ||
| rs7972594 | chr12 | 11724913 | CXCL10 | 3627 | 0.06 | trans | ||
| rs10491979 | chr12 | 11727969 | CXCL10 | 3627 | 0.06 | trans | ||
| rs11171083 | chr12 | 55192688 | CXCL10 | 3627 | 0.08 | trans | ||
| rs1542363 | chr12 | 55213677 | CXCL10 | 3627 | 0.08 | trans | ||
| rs1542364 | chr12 | 55213700 | CXCL10 | 3627 | 0.08 | trans | ||
| rs7298952 | chr12 | 55216414 | CXCL10 | 3627 | 0.08 | trans | ||
| rs921723 | chr12 | 55221358 | CXCL10 | 3627 | 0.08 | trans | ||
| rs2219716 | chr12 | 86139883 | CXCL10 | 3627 | 0.04 | trans | ||
| rs11107750 | chr12 | 95241256 | CXCL10 | 3627 | 0.11 | trans | ||
| rs7304048 | chr12 | 95292947 | CXCL10 | 3627 | 0.11 | trans | ||
| rs4762419 | chr12 | 95771835 | CXCL10 | 3627 | 0.09 | trans | ||
| rs3751152 | chr12 | 121442063 | CXCL10 | 3627 | 0.01 | trans | ||
| rs9645995 | chr13 | 47246048 | CXCL10 | 3627 | 0.01 | trans | ||
| rs9598109 | chr13 | 60878618 | CXCL10 | 3627 | 0.13 | trans | ||
| rs17081091 | chr13 | 66867486 | CXCL10 | 3627 | 0.13 | trans | ||
| rs4328300 | chr13 | 104315373 | CXCL10 | 3627 | 0.12 | trans | ||
| rs9519133 | chr13 | 104325831 | CXCL10 | 3627 | 0.12 | trans | ||
| rs6491786 | chr13 | 104328014 | CXCL10 | 3627 | 0.12 | trans | ||
| rs9519135 | chr13 | 104332307 | CXCL10 | 3627 | 0.12 | trans | ||
| rs9519136 | chr13 | 104332406 | CXCL10 | 3627 | 0.12 | trans | ||
| rs17111522 | chr14 | 41634403 | CXCL10 | 3627 | 0.18 | trans | ||
| rs9989174 | chr14 | 82663493 | CXCL10 | 3627 | 0.02 | trans | ||
| rs1958669 | chr14 | 82675761 | CXCL10 | 3627 | 0.04 | trans | ||
| rs2219618 | chr15 | 54109136 | CXCL10 | 3627 | 0.11 | trans | ||
| rs6494052 | chr15 | 59295924 | CXCL10 | 3627 | 0.11 | trans | ||
| rs6497089 | chr15 | 94163348 | CXCL10 | 3627 | 3.036E-4 | trans | ||
| rs7170017 | chr15 | 94179359 | CXCL10 | 3627 | 0.06 | trans | ||
| rs6498459 | chr16 | 13795898 | CXCL10 | 3627 | 0.07 | trans | ||
| rs963303 | chr16 | 73372497 | CXCL10 | 3627 | 0.01 | trans | ||
| rs4303487 | chr16 | 90033254 | CXCL10 | 3627 | 2.809E-7 | trans | ||
| rs6587203 | chr17 | 19671457 | CXCL10 | 3627 | 0.03 | trans | ||
| rs205113 | chr17 | 19673879 | CXCL10 | 3627 | 0.03 | trans | ||
| rs205111 | chr17 | 19675103 | CXCL10 | 3627 | 0.03 | trans | ||
| rs205110 | chr17 | 19675587 | CXCL10 | 3627 | 0.03 | trans | ||
| rs205093 | chr17 | 19685712 | CXCL10 | 3627 | 0.03 | trans | ||
| rs157406 | chr17 | 19710650 | CXCL10 | 3627 | 0.03 | trans | ||
| rs157402 | chr17 | 19720588 | CXCL10 | 3627 | 0.03 | trans | ||
| rs157396 | chr17 | 19726832 | CXCL10 | 3627 | 0.03 | trans | ||
| rs157397 | chr17 | 19729494 | CXCL10 | 3627 | 0.03 | trans | ||
| rs8074258 | 0 | CXCL10 | 3627 | 0.03 | trans | |||
| rs172186 | chr17 | 19780658 | CXCL10 | 3627 | 0.01 | trans | ||
| rs281334 | chr17 | 19782079 | CXCL10 | 3627 | 0.03 | trans | ||
| rs4586510 | chr17 | 19783973 | CXCL10 | 3627 | 0.03 | trans | ||
| rs2448755 | chr18 | 65344801 | CXCL10 | 3627 | 0.12 | trans | ||
| rs16966813 | chr19 | 32698878 | CXCL10 | 3627 | 4.503E-4 | trans | ||
| rs6076060 | chr20 | 23345068 | CXCL10 | 3627 | 0.12 | trans | ||
| rs965018 | chr20 | 41531585 | CXCL10 | 3627 | 0.18 | trans | ||
| rs9619990 | chr22 | 42163258 | CXCL10 | 3627 | 0.13 | trans | ||
| rs17342476 | chrX | 102243644 | CXCL10 | 3627 | 4.175E-5 | trans | ||
| rs17342483 | chrX | 102250155 | CXCL10 | 3627 | 9.008E-4 | trans | ||
| rs17342504 | chrX | 102272107 | CXCL10 | 3627 | 4.175E-5 | trans | ||
| snp_a-2138851 | 0 | CXCL10 | 3627 | 4.175E-5 | trans | |||
| snp_a-2059928 | 0 | CXCL10 | 3627 | 4.175E-5 | trans | |||
| rs5957194 | chrX | 118769609 | CXCL10 | 3627 | 4.136E-5 | trans |
Section II. Transcriptome annotation
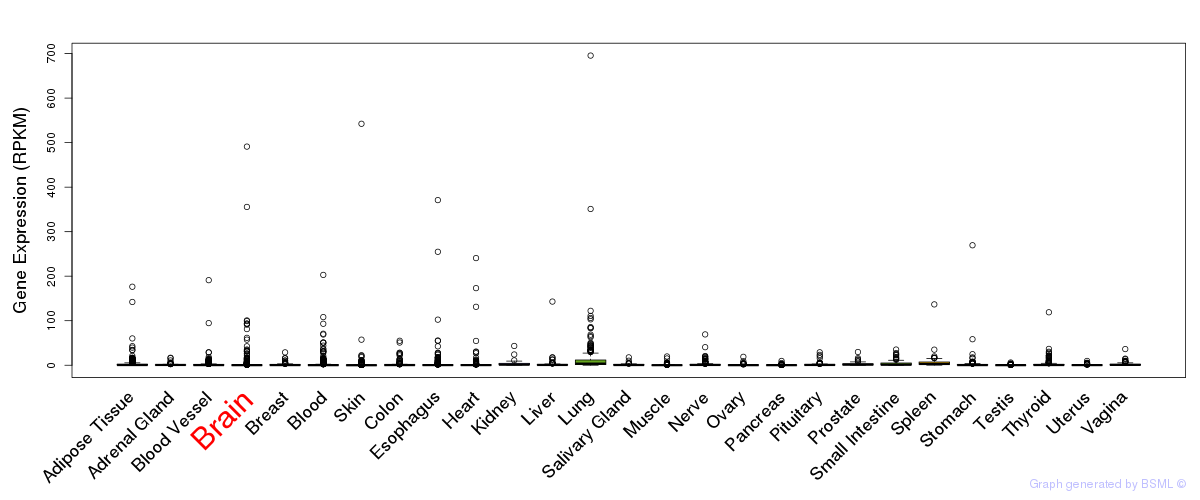
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RFC1 | 0.83 | 0.83 |
| CLPX | 0.83 | 0.84 |
| METT10D | 0.83 | 0.83 |
| HOOK3 | 0.82 | 0.85 |
| ZNF397 | 0.82 | 0.85 |
| ZNF426 | 0.82 | 0.85 |
| TPR | 0.82 | 0.81 |
| XIAP | 0.81 | 0.84 |
| PCM1 | 0.81 | 0.82 |
| NEK4 | 0.81 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.65 | -0.66 |
| AF347015.21 | -0.63 | -0.67 |
| MT-CO2 | -0.61 | -0.63 |
| AF347015.27 | -0.60 | -0.62 |
| HIGD1B | -0.60 | -0.62 |
| IFI27 | -0.60 | -0.62 |
| MT-CYB | -0.58 | -0.60 |
| AF347015.8 | -0.58 | -0.60 |
| AF347015.15 | -0.55 | -0.58 |
| AF347015.33 | -0.54 | -0.56 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| KEGG RIG I LIKE RECEPTOR SIGNALING PATHWAY | 71 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG CYTOSOLIC DNA SENSING PATHWAY | 56 | 44 | All SZGR 2.0 genes in this pathway |
| PID CXCR3 PATHWAY | 43 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE LIGAND BINDING RECEPTORS | 188 | 108 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | 57 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA UP | 205 | 140 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA UP | 294 | 178 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 3D UP | 182 | 110 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D UP | 194 | 122 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP | 204 | 140 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL DIVIDING UP | 57 | 33 | All SZGR 2.0 genes in this pathway |
| GRAHAM NORMAL QUIESCENT VS NORMAL DIVIDING UP | 66 | 47 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| KOINUMA COLON CANCER MSI UP | 16 | 11 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF UP | 215 | 137 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION UP | 211 | 136 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS C UP | 170 | 114 | All SZGR 2.0 genes in this pathway |
| EBAUER TARGETS OF PAX3 FOXO1 FUSION UP | 207 | 128 | All SZGR 2.0 genes in this pathway |
| MAHADEVAN RESPONSE TO MP470 UP | 20 | 11 | All SZGR 2.0 genes in this pathway |
| EBAUER MYOGENIC TARGETS OF PAX3 FOXO1 FUSION | 50 | 26 | All SZGR 2.0 genes in this pathway |
| SEITZ NEOPLASTIC TRANSFORMATION BY 8P DELETION UP | 73 | 47 | All SZGR 2.0 genes in this pathway |
| KANG GIST WITH PDGFRA UP | 50 | 27 | All SZGR 2.0 genes in this pathway |
| YAN ESCAPE FROM ANOIKIS | 24 | 19 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 DN | 156 | 106 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 120 MCF10A | 65 | 44 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP | 223 | 140 | All SZGR 2.0 genes in this pathway |
| LEI HOXC8 TARGETS DN | 17 | 13 | All SZGR 2.0 genes in this pathway |
| RADAEVA RESPONSE TO IFNA1 UP | 52 | 40 | All SZGR 2.0 genes in this pathway |
| WIELAND UP BY HBV INFECTION | 101 | 66 | All SZGR 2.0 genes in this pathway |
| SANA RESPONSE TO IFNG UP | 78 | 50 | All SZGR 2.0 genes in this pathway |
| SANA TNF SIGNALING UP | 83 | 56 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER E2F1 UP | 62 | 35 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP | 183 | 111 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| BROWNE INTERFERON RESPONSIVE GENES | 68 | 44 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 8HR UP | 105 | 73 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL UP | 120 | 89 | All SZGR 2.0 genes in this pathway |
| HAN JNK SINGALING UP | 35 | 21 | All SZGR 2.0 genes in this pathway |
| JACKSON DNMT1 TARGETS UP | 77 | 57 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 8 HR UP | 47 | 39 | All SZGR 2.0 genes in this pathway |
| HILLION HMGA1B TARGETS | 92 | 68 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| MOSERLE IFNA RESPONSE | 31 | 19 | All SZGR 2.0 genes in this pathway |
| GAURNIER PSMD4 TARGETS | 73 | 55 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| PODAR RESPONSE TO ADAPHOSTIN DN | 18 | 16 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| SEKI INFLAMMATORY RESPONSE LPS UP | 77 | 56 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE D7 UP | 107 | 67 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION A | 67 | 52 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE UP | 91 | 63 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP | 181 | 106 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER UP | 288 | 168 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE UP | 109 | 68 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS ICP WITH H3K27ME3 | 54 | 32 | All SZGR 2.0 genes in this pathway |
| KUROZUMI RESPONSE TO ONCOCYTIC VIRUS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| WORSCHECH TUMOR EVASION AND TOLEROGENICITY UP | 30 | 19 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA1 UP | 76 | 46 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP | 217 | 131 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS GROUP2 | 60 | 38 | All SZGR 2.0 genes in this pathway |
| KIM GLIS2 TARGETS UP | 84 | 61 | All SZGR 2.0 genes in this pathway |
| WINZEN DEGRADED VIA KHSRP | 100 | 70 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| BOSCO TH1 CYTOTOXIC MODULE | 114 | 62 | All SZGR 2.0 genes in this pathway |
| ALTEMEIER RESPONSE TO LPS WITH MECHANICAL VENTILATION | 128 | 81 | All SZGR 2.0 genes in this pathway |
| GHANDHI BYSTANDER IRRADIATION UP | 86 | 54 | All SZGR 2.0 genes in this pathway |
| HECKER IFNB1 TARGETS | 95 | 54 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |