Gene Page: EIF3E
Summary ?
| GeneID | 3646 |
| Symbol | EIF3E |
| Synonyms | EIF3-P48|EIF3S6|INT6|eIF3-p46 |
| Description | eukaryotic translation initiation factor 3 subunit E |
| Reference | MIM:602210|HGNC:HGNC:3277|Ensembl:ENSG00000104408|HPRD:03734|Vega:OTTHUMG00000164858 |
| Gene type | protein-coding |
| Map location | 8q22-q23 |
| Pascal p-value | 0.003 |
| Sherlock p-value | 0.105 |
| Fetal beta | 2.122 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0018 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07168148 | 8 | 109260989 | EIF3E | 1.97E-9 | -0.014 | 1.63E-6 | DMG:Jaffe_2016 |
| cg18044723 | 8 | 109261011 | EIF3E | 3.27E-8 | -0.011 | 9.82E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7911247 | chr10 | 49724799 | EIF3E | 3646 | 0.11 | trans |
Section II. Transcriptome annotation
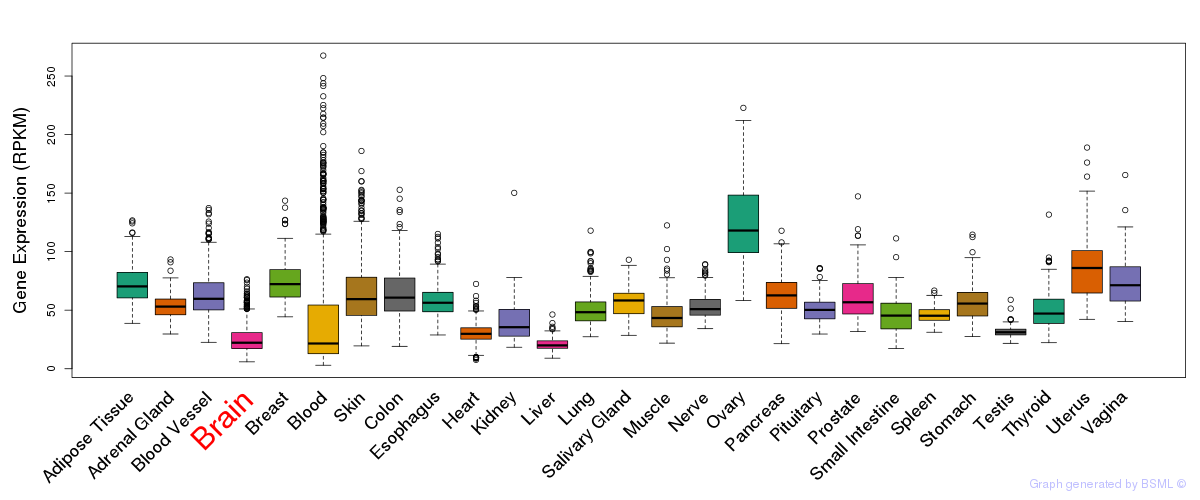
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| COL3A1 | 0.78 | 0.27 |
| LUM | 0.78 | 0.69 |
| C7 | 0.76 | 0.83 |
| ITIH2 | 0.76 | 0.71 |
| OGN | 0.75 | 0.83 |
| OSR1 | 0.75 | 0.66 |
| COL1A1 | 0.74 | 0.50 |
| OLFML3 | 0.74 | 0.54 |
| CYP1B1 | 0.74 | 0.81 |
| COL6A3 | 0.73 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC007216.4 | -0.24 | -0.20 |
| AF347015.21 | -0.24 | 0.05 |
| SNHG12 | -0.24 | -0.47 |
| CHMP4A | -0.23 | -0.33 |
| AC098691.2 | -0.22 | -0.04 |
| AC010300.1 | -0.22 | -0.27 |
| AF347015.18 | -0.22 | -0.03 |
| AC005921.3 | -0.21 | -0.45 |
| MT-CO2 | -0.21 | 0.06 |
| AF347015.26 | -0.21 | 0.09 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003743 | translation initiation factor activity | TAS | 9295280 | |
| GO:0005515 | protein binding | IPI | 14667819 |17220478 |17324924 |17353931 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006446 | regulation of translational initiation | TAS | 9295280 | |
| GO:0006412 | translation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 12588972 | |
| GO:0005852 | eukaryotic translation initiation factor 3 complex | TAS | 9295280 | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BAT2 | D6S51 | D6S51E | DKFZp686D09175 | G2 | HLA-B associated transcript 2 | - | HPRD,BioGRID | 14667819 |
| CNTF | HCNTF | ciliary neurotrophic factor | Affinity Capture-MS | BioGRID | 17353931 |
| COPS6 | CSN6 | MOV34-34KD | COP9 constitutive photomorphogenic homolog subunit 6 (Arabidopsis) | - | HPRD,BioGRID | 12220626 |
| COPS7A | MGC110877 | COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis) | - | HPRD | 12220626 |
| COPS7B | FLJ12612 | MGC111077 | COP9 constitutive photomorphogenic homolog subunit 7B (Arabidopsis) | - | HPRD | 12220626 |
| COPS8 | COP9 | CSN8 | MGC1297 | MGC43256 | SGN8 | COP9 constitutive photomorphogenic homolog subunit 8 (Arabidopsis) | - | HPRD | 12220626 |
| CSN3 | CSN10 | CSNK | KCA | casein kappa | Affinity Capture-Western Two-hybrid | BioGRID | 12220626 |
| DDX24 | - | DEAD (Asp-Glu-Ala-Asp) box polypeptide 24 | Two-hybrid | BioGRID | 16169070 |
| EIF1B | GC20 | eukaryotic translation initiation factor 1B | Affinity Capture-MS | BioGRID | 17353931 |
| EIF3A | EIF3 | EIF3S10 | KIAA0139 | P167 | TIF32 | eIF3-p170 | eIF3-theta | p180 | p185 | eukaryotic translation initiation factor 3, subunit A | Affinity Capture-MS Affinity Capture-Western Co-purification | BioGRID | 11590142 |14519125 |17353931 |
| EIF3C | EIF3S8 | FLJ78287 | MGC189744 | eIF3-p110 | eukaryotic translation initiation factor 3, subunit C | - | HPRD,BioGRID | 10504338 |
| EIF3EIP | EIF3S11 | EIF3S6IP | HSPC021 | HSPC025 | MSTP005 | eukaryotic translation initiation factor 3, subunit E interacting protein | - | HPRD,BioGRID | 11590142 |
| GPAA1 | GAA1 | hGAA1 | glycosylphosphatidylinositol anchor attachment protein 1 homolog (yeast) | Two-hybrid | BioGRID | 16169070 |
| GPBP1L1 | RP11-767N6.1 | SP192 | GC-rich promoter binding protein 1-like 1 | Two-hybrid | BioGRID | 16189514 |
| HAP1 | HAP2 | HIP5 | HLP | hHLP1 | huntingtin-associated protein 1 | Two-hybrid | BioGRID | 16169070 |
| IFIT1 | G10P1 | GARG-16 | IFI-56 | IFI56 | IFNAI1 | ISG56 | RNM561 | interferon-induced protein with tetratricopeptide repeats 1 | - | HPRD,BioGRID | 10644362 |
| MAGED1 | DLXIN-1 | NRAGE | melanoma antigen family D, 1 | Affinity Capture-MS | BioGRID | 17353931 |
| NSF | SKD2 | N-ethylmaleimide-sensitive factor | Two-hybrid | BioGRID | 16169070 |
| RGS2 | G0S8 | regulator of G-protein signaling 2, 24kDa | Affinity Capture-MS | BioGRID | 17353931 |
| RUNDC3A | RAP2IP | RPIP8 | RUN domain containing 3A | Two-hybrid | BioGRID | 16189514 |
| SAP18 | 2HOR0202 | MGC27131 | SAP18p | Sin3A-associated protein, 18kDa | Affinity Capture-MS | BioGRID | 17353931 |
| TRIM27 | RFP | RNF76 | tripartite motif-containing 27 | - | HPRD | 10504338 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID HIF2PATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSLATION | 222 | 75 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | 74 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | 84 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | 176 | 51 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| DER IFN GAMMA RESPONSE DN | 11 | 9 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN | 141 | 92 | All SZGR 2.0 genes in this pathway |
| KIM LRRC3B TARGETS | 30 | 24 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| NUTT GBM VS AO GLIOMA DN | 45 | 22 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS ONCOGENIC SIGNATURE | 89 | 56 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA | 43 | 27 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS UP | 74 | 45 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE DN | 181 | 97 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 9 | 76 | 45 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM SENSITIVE GENES | 90 | 54 | All SZGR 2.0 genes in this pathway |
| BILANGES RAPAMYCIN SENSITIVE VIA TSC1 AND TSC2 | 73 | 37 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |