Gene Page: ITGA3
Summary ?
| GeneID | 3675 |
| Symbol | ITGA3 |
| Synonyms | CD49C|FRP-2|GAP-B3|GAPB3|ILNEB|MSK18|VCA-2|VL3A|VLA3a |
| Description | integrin subunit alpha 3 |
| Reference | MIM:605025|HGNC:HGNC:6139|Ensembl:ENSG00000005884|HPRD:05431|Vega:OTTHUMG00000161890 |
| Gene type | protein-coding |
| Map location | 17q21.33 |
| Pascal p-value | 0.581 |
| Sherlock p-value | 0.127 |
| Fetal beta | -0.374 |
| eGene | Cerebellar Hemisphere Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Gulsuner_2013 | Whole Exome Sequencing analysis | 155 DNMs identified by exome sequencing of quads or trios of schizophrenia individuals and their parents. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ITGA3 | chr17 | 48154453 | C | G | NM_002204 NM_005501 | p.680P>A p.680P>A | missense missense | Schizophrenia | DNM:Gulsuner_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1570589 | chr9 | 116874354 | ITGA3 | 3675 | 0.15 | trans |
Section II. Transcriptome annotation
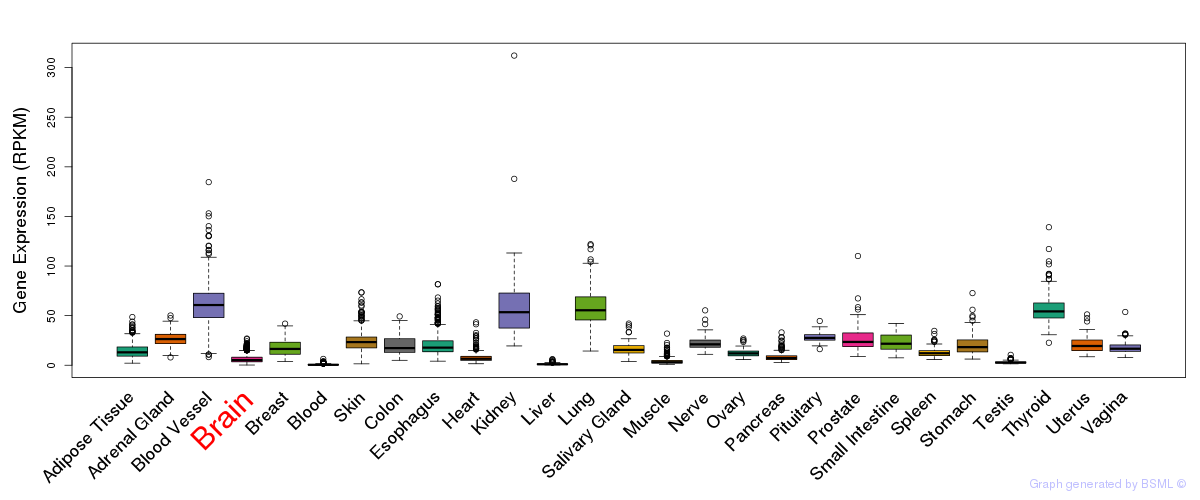
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ATHL1 | 0.72 | 0.78 |
| ADAM33 | 0.72 | 0.77 |
| ACADVL | 0.72 | 0.73 |
| MYO15B | 0.69 | 0.81 |
| CATSPER2 | 0.69 | 0.70 |
| DNAH1 | 0.69 | 0.77 |
| AL117190.3 | 0.68 | 0.81 |
| DCDC2B | 0.67 | 0.69 |
| CES8 | 0.67 | 0.64 |
| MALAT1 | 0.65 | 0.69 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MOBKL3 | -0.47 | -0.56 |
| HSPA13 | -0.45 | -0.49 |
| DNAJA1 | -0.44 | -0.51 |
| METTL9 | -0.44 | -0.50 |
| ARF4 | -0.44 | -0.48 |
| SERP1 | -0.44 | -0.50 |
| UBE2N | -0.44 | -0.56 |
| RBMXL1 | -0.44 | -0.45 |
| MARCKS | -0.44 | -0.43 |
| IPMK | -0.44 | -0.47 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 14676841 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001764 | neuron migration | IEA | neuron (GO term level: 8) | - |
| GO:0007160 | cell-matrix adhesion | TAS | 1655803 | |
| GO:0007155 | cell adhesion | IEA | - | |
| GO:0007229 | integrin-mediated signaling pathway | IEA | - | |
| GO:0007613 | memory | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0019717 | synaptosome | IEA | Synap, Brain (GO term level: 7) | - |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0008305 | integrin complex | IEA | - | |
| GO:0008305 | integrin complex | TAS | 1655803 | |
| GO:0016323 | basolateral plasma membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BIN1 | AMPH2 | AMPHL | DKFZp547F068 | MGC10367 | SH3P9 | bridging integrator 1 | - | HPRD,BioGRID | 10094488 |
| CALR | CRT | FLJ26680 | RO | SSA | cC1qR | calreticulin | - | HPRD | 8006073|11327697 |
| CALR | CRT | FLJ26680 | RO | SSA | cC1qR | calreticulin | - | HPRD | 11327697 |
| CD151 | GP27 | MER2 | PETA-3 | RAPH | SFA1 | TSPAN24 | CD151 molecule (Raph blood group) | Reconstituted Complex | BioGRID | 10734060 |
| CD82 | 4F9 | C33 | GR15 | IA4 | KAI1 | R2 | SAR2 | ST6 | TSPAN27 | CD82 molecule | - | HPRD | 8757325 |
| CD9 | 5H9 | BA2 | BTCC-1 | DRAP-27 | GIG2 | MIC3 | MRP-1 | P24 | TSPAN29 | CD9 molecule | - | HPRD,BioGRID | 10065872 |10694273 |
| FHL2 | AAG11 | DRAL | SLIM3 | four and a half LIM domains 2 | - | HPRD,BioGRID | 10906324 |
| FN1 | CIG | DKFZp686F10164 | DKFZp686H0342 | DKFZp686I1370 | DKFZp686O13149 | ED-B | FINC | FN | FNZ | GFND | GFND2 | LETS | MSF | fibronectin 1 | - | HPRD | 9733622 |
| ITGB1 | CD29 | FNRB | GPIIA | MDF2 | MSK12 | VLA-BETA | VLAB | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | VLA3-alpha3 interacts with VLA3-beta. | BIND | 1690718 |1693624 |3546305 |
| ITGB1 | CD29 | FNRB | GPIIA | MDF2 | MSK12 | VLA-BETA | VLAB | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | VLA3-alpha3 interacts with VLA3-beta | BIND | 3546305 |
| ITGB1 | CD29 | FNRB | GPIIA | MDF2 | MSK12 | VLA-BETA | VLAB | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | - | HPRD,BioGRID | 15254262 |
| LAMA3 | BM600 | E170 | LAMNA | LOCS | lama3a | laminin, alpha 3 | - | HPRD | 9950675 |11278628 |
| LGALS8 | Gal-8 | PCTA-1 | PCTA1 | Po66-CBP | lectin, galactoside-binding, soluble, 8 | - | HPRD,BioGRID | 10852818 |
| RABIF | MSS4 | RASGFR3 | RASGRF3 | RAB interacting factor | - | HPRD,BioGRID | 10094488 |
| TIMP2 | CSC-21K | TIMP metallopeptidase inhibitor 2 | TIMP2 interacts with ITGA3 (alpha3). | BIND | 12887919 |
| TSPAN4 | NAG-2 | NAG2 | TETRASPAN | TM4SF7 | TSPAN-4 | tetraspanin 4 | - | HPRD,BioGRID | 9360996 |10734060 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ECM RECEPTOR INTERACTION | 84 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG HEMATOPOIETIC CELL LINEAGE | 88 | 60 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| KEGG HYPERTROPHIC CARDIOMYOPATHY HCM | 85 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG ARRHYTHMOGENIC RIGHT VENTRICULAR CARDIOMYOPATHY ARVC | 76 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG DILATED CARDIOMYOPATHY | 92 | 68 | All SZGR 2.0 genes in this pathway |
| ST INTEGRIN SIGNALING PATHWAY | 82 | 62 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN1 PATHWAY | 66 | 44 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN CS PATHWAY | 26 | 16 | All SZGR 2.0 genes in this pathway |
| PID ARF6 TRAFFICKING PATHWAY | 49 | 34 | All SZGR 2.0 genes in this pathway |
| PID REELIN PATHWAY | 29 | 29 | All SZGR 2.0 genes in this pathway |
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| PID UPA UPAR PATHWAY | 42 | 30 | All SZGR 2.0 genes in this pathway |
| PID IL23 PATHWAY | 37 | 30 | All SZGR 2.0 genes in this pathway |
| PID DELTA NP63 PATHWAY | 47 | 34 | All SZGR 2.0 genes in this pathway |
| PID TAP63 PATHWAY | 54 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | 91 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME BASIGIN INTERACTIONS | 30 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN CELL SURFACE INTERACTIONS | 79 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| CORRE MULTIPLE MYELOMA UP | 74 | 45 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| WANG BARRETTS ESOPHAGUS UP | 51 | 27 | All SZGR 2.0 genes in this pathway |
| BEGUM TARGETS OF PAX3 FOXO1 FUSION DN | 45 | 34 | All SZGR 2.0 genes in this pathway |
| MAHADEVAN GIST MORPHOLOGICAL SWITCH | 15 | 11 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| COLIN PILOCYTIC ASTROCYTOMA VS GLIOBLASTOMA UP | 35 | 32 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| CROMER METASTASIS DN | 81 | 58 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C8 | 72 | 56 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS UP | 56 | 35 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA UP | 52 | 33 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR DN | 101 | 70 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 8HR DN | 47 | 31 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C1 | 19 | 15 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA EARLY RESPONSE TO TGFB1 | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN WITHOUT MGMT 48HR DN | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| HELLEBREKERS SILENCED DURING TUMOR ANGIOGENESIS | 80 | 56 | All SZGR 2.0 genes in this pathway |
| GU PDEF TARGETS UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER DN | 134 | 83 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K4ME3 AND H3K27ME3 | 210 | 148 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS EARLY DN | 38 | 27 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS LATE DN | 38 | 25 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS DN | 87 | 66 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN DN | 88 | 68 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 2HR DN | 49 | 33 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| VANDESLUIS COMMD1 TARGETS GROUP 3 UP | 89 | 50 | All SZGR 2.0 genes in this pathway |
| WIEDERSCHAIN TARGETS OF BMI1 AND PCGF2 | 57 | 39 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP | 169 | 105 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124/506 | 1160 | 1166 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-181 | 883 | 890 | 1A,m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-24 | 410 | 416 | 1A | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-381 | 1169 | 1175 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.