Gene Page: JAK2
Summary ?
| GeneID | 3717 |
| Symbol | JAK2 |
| Synonyms | JTK10|THCYT3 |
| Description | Janus kinase 2 |
| Reference | MIM:147796|HGNC:HGNC:6192|Ensembl:ENSG00000096968|HPRD:00993|Vega:OTTHUMG00000019490 |
| Gene type | protein-coding |
| Map location | 9p24 |
| Pascal p-value | 0.16 |
| Fetal beta | 0.202 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 4.0087 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
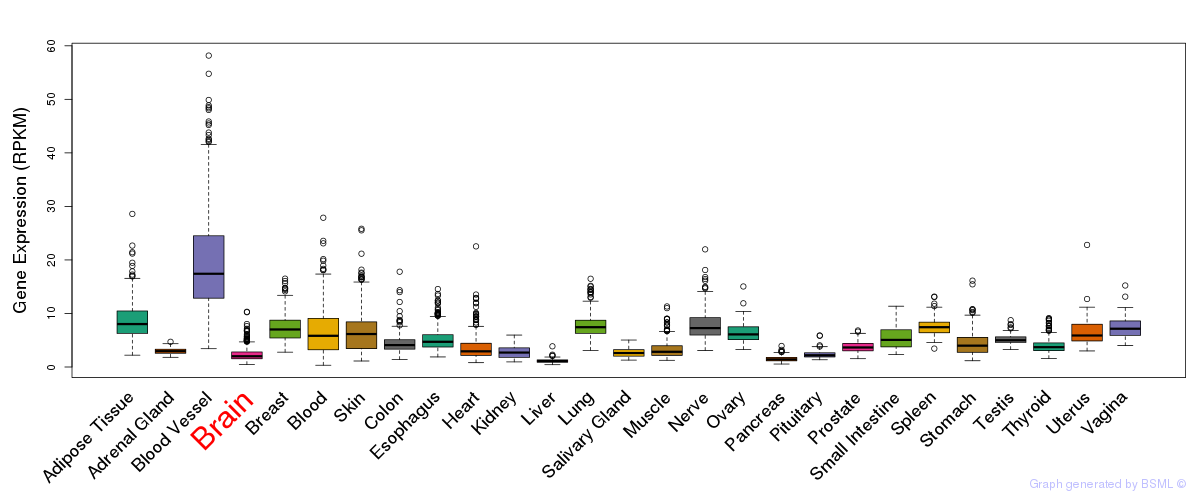
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIF3C | 0.90 | 0.94 |
| CEP68 | 0.89 | 0.92 |
| INPP4A | 0.89 | 0.92 |
| CECR6 | 0.88 | 0.91 |
| ATCAY | 0.88 | 0.92 |
| CAPN5 | 0.88 | 0.92 |
| KLHL18 | 0.88 | 0.93 |
| KIAA0317 | 0.88 | 0.93 |
| DCLK1 | 0.88 | 0.91 |
| MAPT | 0.88 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.70 | -0.79 |
| SERPINB6 | -0.69 | -0.75 |
| AF347015.31 | -0.68 | -0.79 |
| S100B | -0.68 | -0.77 |
| AC021016.1 | -0.68 | -0.79 |
| HEPN1 | -0.68 | -0.71 |
| MT-CO2 | -0.68 | -0.80 |
| AF347015.33 | -0.67 | -0.76 |
| COPZ2 | -0.67 | -0.74 |
| TSC22D4 | -0.67 | -0.72 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005102 | receptor binding | IPI | Neurotransmitter (GO term level: 4) | 11201744 |
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004718 | Janus kinase activity | IEA | - | |
| GO:0004713 | protein tyrosine kinase activity | IEA | - | |
| GO:0004715 | non-membrane spanning protein tyrosine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0042169 | SH2 domain binding | IPI | 9727029 | |
| GO:0016301 | kinase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0030099 | myeloid cell differentiation | IEA | - | |
| GO:0007262 | STAT protein nuclear translocation | IEA | - | |
| GO:0007260 | tyrosine phosphorylation of STAT protein | IEA | - | |
| GO:0007167 | enzyme linked receptor protein signaling pathway | IEA | - | |
| GO:0008285 | negative regulation of cell proliferation | IEA | - | |
| GO:0007498 | mesoderm development | TAS | 9590174 | |
| GO:0006928 | cell motion | TAS | 9670957 | |
| GO:0006915 | apoptosis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0012505 | endomembrane system | IEA | - | |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0045121 | membrane raft | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AGTR1 | AG2S | AGTR1A | AGTR1B | AT1 | AT1B | AT1R | AT2R1 | AT2R1A | AT2R1B | HAT1R | angiotensin II receptor, type 1 | - | HPRD | 7746328 |
| BCR | ALL | BCR-ABL1 | BCR1 | CML | D22S11 | D22S662 | FLJ16453 | PHL | breakpoint cluster region | - | HPRD,BioGRID | 12023981 |12370803 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD,BioGRID | 11163768 |
| CCR2 | CC-CKR-2 | CCR2A | CCR2B | CD192 | CKR2 | CKR2A | CKR2B | CMKBR2 | MCP-1-R | chemokine (C-C motif) receptor 2 | - | HPRD,BioGRID | 9670957 |
| CCR5 | CC-CKR-5 | CCCKR5 | CD195 | CKR-5 | CKR5 | CMKBR5 | chemokine (C-C motif) receptor 5 | - | HPRD | 11591716 |
| CCR5 | CC-CKR-5 | CCCKR5 | CD195 | CKR-5 | CKR5 | CMKBR5 | chemokine (C-C motif) receptor 5 | Affinity Capture-Western | BioGRID | 11278738 |
| CSF2RB | CD131 | CDw131 | IL3RB | IL5RB | colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) | Affinity Capture-Western in vivo Reconstituted Complex | BioGRID | 7527392 |7775438 |10772872 |
| CSF3R | CD114 | GCSFR | colony stimulating factor 3 receptor (granulocyte) | - | HPRD | 7775438 |
| CTLA4 | CD152 | CELIAC3 | CTLA-4 | GSE | IDDM12 | cytotoxic T-lymphocyte-associated protein 4 | CTLA-4 interacts with Jak2. | BIND | 10842319 |
| CTLA4 | CD152 | CELIAC3 | CTLA-4 | GSE | IDDM12 | cytotoxic T-lymphocyte-associated protein 4 | - | HPRD,BioGRID | 10842319 |
| CXCR4 | CD184 | D2S201E | FB22 | HM89 | HSY3RR | LAP3 | LCR1 | LESTR | NPY3R | NPYR | NPYRL | NPYY3R | WHIM | chemokine (C-X-C motif) receptor 4 | - | HPRD,BioGRID | 10506573 |
| DNAJA3 | FLJ45758 | TID1 | hTid-1 | DnaJ (Hsp40) homolog, subfamily A, member 3 | - | HPRD,BioGRID | 11679576 |
| DNAJA3 | FLJ45758 | TID1 | hTid-1 | DnaJ (Hsp40) homolog, subfamily A, member 3 | JAK2 interacts with Tid-1S. | BIND | 11679576 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | Affinity Capture-Western | BioGRID | 10358079 |
| ELP2 | FLJ10879 | SHINC-2 | STATIP1 | StIP | elongation protein 2 homolog (S. cerevisiae) | - | HPRD,BioGRID | 10954736 |
| EPOR | MGC138358 | erythropoietin receptor | - | HPRD,BioGRID | 8343951 |11779507 |
| ERBB2 | CD340 | HER-2 | HER-2/neu | HER2 | NEU | NGL | TKR1 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | - | HPRD,BioGRID | 11940572 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 10551884 |
| GHR | GHBP | growth hormone receptor | - | HPRD,BioGRID | 7540178 |8063815 |10502458 |
| GRB10 | GRB-IR | Grb-10 | IRBP | KIAA0207 | MEG1 | RSS | growth factor receptor-bound protein 10 | - | HPRD,BioGRID | 9632636 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 7500025 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | - | HPRD,BioGRID | 11679576 |
| HTR2A | 5-HT2A | HTR2 | 5-hydroxytryptamine (serotonin) receptor 2A | - | HPRD,BioGRID | 9169451 |
| IFNGR1 | CD119 | FLJ45734 | IFNGR | interferon gamma receptor 1 | Affinity Capture-Western | BioGRID | 7673114 |
| IFNGR2 | AF-1 | IFGR2 | IFNGT1 | interferon gamma receptor 2 (interferon gamma transducer 1) | - | HPRD | 7514165 |
| IFNGR2 | AF-1 | IFGR2 | IFNGT1 | interferon gamma receptor 2 (interferon gamma transducer 1) | Reconstituted Complex | BioGRID | 7673114 |
| IL12RB2 | - | interleukin 12 receptor, beta 2 | JAK2 interacts with IL12RB2. | BIND | 9038232 |
| IL12RB2 | - | interleukin 12 receptor, beta 2 | - | HPRD,BioGRID | 10198225 |
| IL23R | - | interleukin 23 receptor | - | HPRD | 12023369 |
| IL5RA | CD125 | CDw125 | HSIL5R3 | IL5R | MGC26560 | interleukin 5 receptor, alpha | - | HPRD,BioGRID | 9516124 |
| IL6ST | CD130 | CDw130 | GP130 | GP130-RAPS | IL6R-beta | interleukin 6 signal transducer (gp130, oncostatin M receptor) | Affinity Capture-Western | BioGRID | 9388212 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | Affinity Capture-Western Biochemical Activity | BioGRID | 9492017 |11208867 |
| KIT | C-Kit | CD117 | PBT | SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | - | HPRD,BioGRID | 7527392 |8611693 |9355737 |
| LEPR | CD295 | OBR | leptin receptor | - | HPRD | 8608603 |
| LYN | FLJ26625 | JTK8 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | - | HPRD,BioGRID | 9573010 |
| MDK | FLJ27379 | MK | NEGF2 | midkine (neurite growth-promoting factor 2) | - | HPRD | 9452495 |
| OSMR | MGC150626 | MGC150627 | MGC75127 | OSMRB | oncostatin M receptor | - | HPRD | 10586060 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | - | HPRD,BioGRID | 8702385 |
| PPIA | CYPA | CYPH | MGC117158 | MGC12404 | MGC23397 | peptidylprolyl isomerase A (cyclophilin A) | - | HPRD,BioGRID | 12668872 |
| PPP2R1B | MGC26454 | PR65B | protein phosphatase 2 (formerly 2A), regulatory subunit A, beta isoform | - | HPRD,BioGRID | 11440634 |
| PPP2R4 | MGC2184 | PP2A | PR53 | PTPA | protein phosphatase 2A activator, regulatory subunit 4 | Affinity Capture-Western | BioGRID | 8702385 |
| PPP2R5A | B56A | MGC131915 | PR61A | protein phosphatase 2, regulatory subunit B', alpha isoform | - | HPRD | 8702385 |
| PRLR | hPRLrI | prolactin receptor | - | HPRD,BioGRID | 10772872 |
| PRMT5 | HRMT1L5 | IBP72 | JBP1 | SKB1 | SKB1Hs | protein arginine methyltransferase 5 | - | HPRD,BioGRID | 10531356 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD,BioGRID | 9553131 |10925297 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD,BioGRID | 11818507 |
| PTPN1 | PTP1B | protein tyrosine phosphatase, non-receptor type 1 | - | HPRD,BioGRID | 11694501 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | Biochemical Activity in vivo Reconstituted Complex | BioGRID | 8639815 |8912646 |8995399 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | - | HPRD | 7559603 |8912646 |8995399 |
| PTPN12 | PTP-PEST | PTPG1 | protein tyrosine phosphatase, non-receptor type 12 | - | HPRD,BioGRID | 11731619 |
| PTPN6 | HCP | HCPH | HPTP1C | PTP-1C | SH-PTP1 | SHP-1 | SHP-1L | SHP1 | protein tyrosine phosphatase, non-receptor type 6 | - | HPRD,BioGRID | 8943354 |
| PTPRC | B220 | CD45 | CD45R | GP180 | LCA | LY5 | T200 | protein tyrosine phosphatase, receptor type, C | - | HPRD | 11201744 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | - | HPRD,BioGRID | 8876196 |
| SH2B1 | DKFZp547G1110 | FLJ30542 | KIAA1299 | SH2-B | SH2B | SH2B adaptor protein 1 | - | HPRD,BioGRID | 9343427 |
| SH2B1 | DKFZp547G1110 | FLJ30542 | KIAA1299 | SH2-B | SH2B | SH2B adaptor protein 1 | - | HPRD | 10757801 |
| SH2B2 | APS | SH2B adaptor protein 2 | Biochemical Activity | BioGRID | 10374881 |
| SH2B2 | APS | SH2B adaptor protein 2 | - | HPRD | 10872802 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD,BioGRID | 7535773 |9126968 |
| SIRPA | BIT | CD172A | MFR | MYD-1 | P84 | PTPNS1 | SHPS-1 | SHPS1 | SIRP | SIRP-ALPHA-1 | SIRPalpha | SIRPalpha2 | signal-regulatory protein alpha | - | HPRD,BioGRID | 10842184 |
| SOCS1 | CIS1 | CISH1 | JAB | SOCS-1 | SSI-1 | SSI1 | TIP3 | suppressor of cytokine signaling 1 | - | HPRD | 9202126 |10064597 |11971965 |
| SOCS1 | CIS1 | CISH1 | JAB | SOCS-1 | SSI-1 | SSI1 | TIP3 | suppressor of cytokine signaling 1 | Affinity Capture-Western in vitro in vivo Reconstituted Complex Two-hybrid | BioGRID | 9202126 |10064597 |10455112 |11713228 |11971965 |
| SOCS3 | ATOD4 | CIS3 | Cish3 | MGC71791 | SOCS-3 | SSI-3 | SSI3 | suppressor of cytokine signaling 3 | - | HPRD,BioGRID | 9344848 |10421843 |
| STAM | DKFZp686J2352 | STAM1 | signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 | - | HPRD,BioGRID | 9133424 |
| STAM2 | DKFZp564C047 | Hbp | STAM2A | STAM2B | signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 | - | HPRD,BioGRID | 10899310 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | - | HPRD | 10925297 |
| STAT5A | MGF | STAT5 | signal transducer and activator of transcription 5A | - | HPRD,BioGRID | 9575217 |
| STAT5B | STAT5 | signal transducer and activator of transcription 5B | Affinity Capture-Western Two-hybrid | BioGRID | 9047382 |9575217 |
| TEC | MGC126760 | MGC126762 | PSCTK4 | tec protein tyrosine kinase | - | HPRD,BioGRID | 9473212 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | - | HPRD,BioGRID | 9510175 |
| TSHR | CHNG1 | LGR3 | MGC75129 | hTSHR-I | thyroid stimulating hormone receptor | - | HPRD,BioGRID | 10809230 |
| TUB | rd5 | tubby homolog (mouse) | - | HPRD,BioGRID | 10455176 |
| UBASH3B | KIAA1959 | MGC15437 | STS-1 | STS1 | p70 | ubiquitin associated and SH3 domain containing, B | - | HPRD | 12370296 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | - | HPRD,BioGRID | 7530656 |9162069 |
| VCP | IBMPFD | MGC131997 | MGC148092 | MGC8560 | TERA | p97 | valosin-containing protein | - | HPRD,BioGRID | 11087817 |
| YES1 | HsT441 | P61-YES | Yes | c-yes | v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 | - | HPRD,BioGRID | 8702385 |
Section V. Pathway annotation
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-135 | 935 | 942 | 1A,m8 | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-216 | 833 | 840 | 1A,m8 | hsa-miR-216 | UAAUCUCAGCUGGCAACUGUG |
| miR-375 | 636 | 642 | 1A | hsa-miR-375 | UUUGUUCGUUCGGCUCGCGUGA |
| miR-381 | 656 | 662 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-433-3p | 101 | 107 | m8 | hsa-miR-433brain | AUCAUGAUGGGCUCCUCGGUGU |
| miR-496 | 325 | 331 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.