Gene Page: IPO5
Summary ?
| GeneID | 3843 |
| Symbol | IPO5 |
| Synonyms | IMB3|KPNB3|Pse1|RANBP5|imp5 |
| Description | importin 5 |
| Reference | MIM:602008|HGNC:HGNC:6402|Ensembl:ENSG00000065150|HPRD:03597|Vega:OTTHUMG00000017244 |
| Gene type | protein-coding |
| Map location | 13q32.2 |
| Pascal p-value | 0.939 |
| Sherlock p-value | 0.005 |
| Fetal beta | 0.421 |
| DMG | 1 (# studies) |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Expression | Meta-analysis of gene expression | P value: 1.509 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.2023 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg05126147 | 13 | 98628060 | IPO5 | 3.42E-10 | -0.008 | 7.22E-7 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
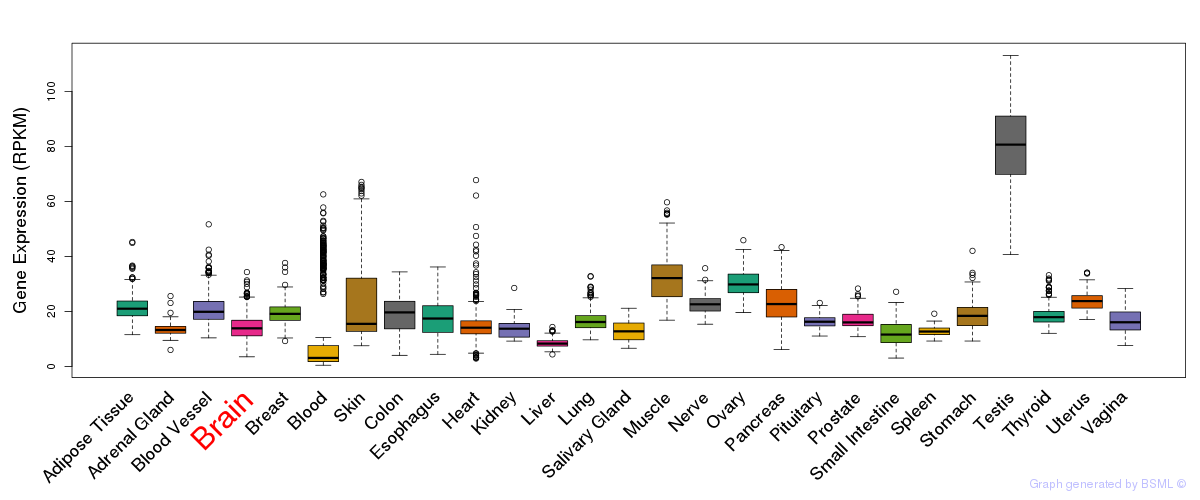
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005095 | GTPase inhibitor activity | TAS | 9271386 | |
| GO:0005488 | binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 15282309 | |
| GO:0008536 | Ran GTPase binding | TAS | 9114010 | |
| GO:0008565 | protein transporter activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000059 | protein import into nucleus, docking | IEA | - | |
| GO:0006607 | NLS-bearing substrate import into nucleus | TAS | 9114010 | |
| GO:0006886 | intracellular protein transport | IEA | - | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | TAS | 9114010 | |
| GO:0005643 | nuclear pore | TAS | 9271386 | |
| GO:0005737 | cytoplasm | TAS | 9114010 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| H1F0 | H10 | H1FV | MGC5241 | H1 histone family, member 0 | in vitro | BioGRID | 10228156 |
| NUP153 | HNUP153 | N153 | nucleoporin 153kDa | - | HPRD,BioGRID | 9114010 |
| NUP214 | CAIN | CAN | D9S46E | MGC104525 | N214 | nucleoporin 214kDa | - | HPRD,BioGRID | 9114010 |
| NUP98 | ADIR2 | NUP196 | NUP96 | nucleoporin 98kDa | - | HPRD | 9114010 |
| RAG2 | RAG-2 | recombination activating gene 2 | - | HPRD | 12861017 |
| RAN | ARA24 | Gsp1 | TC4 | RAN, member RAS oncogene family | - | HPRD,BioGRID | 9114010 |
| RANBP2 | NUP358 | TRP1 | TRP2 | RAN binding protein 2 | - | HPRD | 9114010 |
| RPL23A | FLJ27455 | MDA20 | ribosomal protein L23a | Affinity Capture-Western in vitro | BioGRID | 9687515 |11682607 |
| RPL5 | MGC117339 | MSTP030 | ribosomal protein L5 | - | HPRD,BioGRID | 9687515 |
| RPS7 | - | ribosomal protein S7 | - | HPRD,BioGRID | 9687515 |
| SRP19 | - | signal recognition particle 19kDa | Affinity Capture-Western Reconstituted Complex | BioGRID | 11682607 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME INFLUENZA LIFE CYCLE | 203 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | 169 | 47 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA UP | 294 | 178 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS UP | 194 | 112 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS REPRESSED BY SERUM | 159 | 93 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED MODERATELY VS POORLY UP | 121 | 71 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| KAMMINGA EZH2 TARGETS | 41 | 26 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS DN | 215 | 132 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL SHORT TERM | 32 | 15 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 24HR | 43 | 30 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN | 202 | 132 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| TOOKER GEMCITABINE RESISTANCE DN | 122 | 84 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE UP | 163 | 102 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| IRITANI MAD1 TARGETS DN | 47 | 30 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 UP | 188 | 121 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 14 | 143 | 86 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS UP | 127 | 78 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 4 | 10 | 1A | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-181 | 588 | 594 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-186 | 1248 | 1254 | 1A | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-23 | 590 | 597 | 1A,m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-323 | 590 | 596 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-377 | 1059 | 1066 | 1A,m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-505 | 1145 | 1151 | 1A | hsa-miR-505 | GUCAACACUUGCUGGUUUCCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.