Gene Page: RHOA
Summary ?
| GeneID | 387 |
| Symbol | RHOA |
| Synonyms | ARH12|ARHA|RHO12|RHOH12 |
| Description | ras homolog family member A |
| Reference | MIM:165390|HGNC:HGNC:667|Ensembl:ENSG00000067560|HPRD:01323|Vega:OTTHUMG00000156838 |
| Gene type | protein-coding |
| Map location | 3p21.3 |
| Pascal p-value | 0.026 |
| Sherlock p-value | 0.006 |
| Fetal beta | -0.426 |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0603 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg16951854 | 3 | 49449507 | RHOA;TCTA | 9.37E-5 | 0.351 | 0.027 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
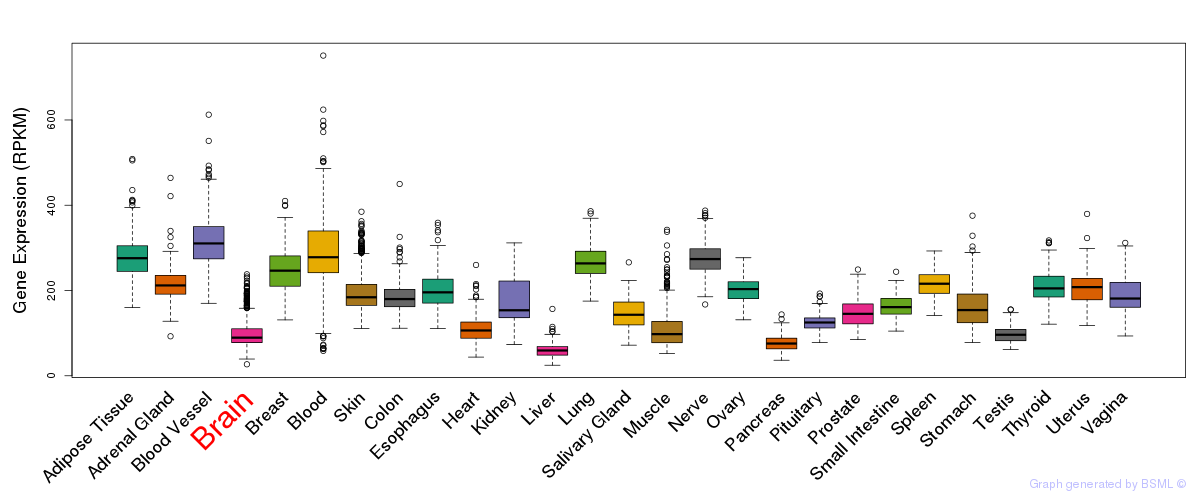
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| H2AFY2 | 0.87 | 0.92 |
| STAG1 | 0.86 | 0.89 |
| PHF16 | 0.85 | 0.81 |
| RNF122 | 0.85 | 0.72 |
| LRCH2 | 0.84 | 0.92 |
| KDM1 | 0.84 | 0.94 |
| ZNF491 | 0.84 | 0.79 |
| ZNF738 | 0.84 | 0.85 |
| ZBTB5 | 0.84 | 0.90 |
| PRIM1 | 0.84 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.60 | -0.84 |
| TNFSF12 | -0.59 | -0.67 |
| C5orf53 | -0.59 | -0.78 |
| PTH1R | -0.58 | -0.74 |
| FBXO2 | -0.58 | -0.68 |
| AIFM3 | -0.58 | -0.81 |
| ALDOC | -0.58 | -0.74 |
| LHPP | -0.57 | -0.64 |
| LDHD | -0.56 | -0.69 |
| SLC9A3R2 | -0.56 | -0.57 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0003924 | GTPase activity | TAS | 10436159 | |
| GO:0005515 | protein binding | IPI | 15644318 | |
| GO:0005525 | GTP binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007519 | skeletal muscle development | IEA | neuron (GO term level: 8) | - |
| GO:0043524 | negative regulation of neuron apoptosis | IEA | neuron (GO term level: 9) | - |
| GO:0000902 | cell morphogenesis | IEA | - | |
| GO:0007160 | cell-matrix adhesion | IEA | - | |
| GO:0007155 | cell adhesion | IEA | - | |
| GO:0007266 | Rho protein signal transduction | TAS | 10436159 | |
| GO:0007264 | small GTPase mediated signal transduction | IEA | - | |
| GO:0043149 | stress fiber formation | IEA | - | |
| GO:0042346 | positive regulation of NF-kappaB import into nucleus | NAS | 12761501 | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0030036 | actin cytoskeleton organization | TAS | 10436159 | |
| GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB cascade | IEP | 12761501 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | IEA | - | |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKAP13 | AKAP-Lbc | BRX | FLJ11952 | FLJ43341 | HA-3 | Ht31 | LBC | PROTO-LB | PROTO-LBC | c-lbc | A kinase (PRKA) anchor protein 13 | - | HPRD,BioGRID | 11696353 |12423633 |
| ARHGAP1 | CDC42GAP | RHOGAP | RHOGAP1 | p50rhoGAP | Rho GTPase activating protein 1 | - | HPRD,BioGRID | 9407060 |9548756 |
| ARHGAP10 | FLJ20896 | FLJ41791 | GRAF2 | PS-GAP | Rho GTPase activating protein 10 | Biochemical Activity | BioGRID | 11432776 |
| ARHGAP21 | ARHGAP10 | DKFZp761L0424 | FLJ33323 | FLJ90108 | Rho GTPase activating protein 21 | ARHGAP21 interacts with RhoA-GTP, and acts as a GAP to increase RhoA GTPase activity. | BIND | 15793564 |
| ARHGAP26 | FLJ42530 | GRAF | KIAA0621 | OPHN1L | OPHN1L1 | Rho GTPase activating protein 26 | Biochemical Activity | BioGRID | 11432776 |
| ARHGAP5 | GFI2 | RhoGAP5 | p190-B | Rho GTPase activating protein 5 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12842009 |
| ARHGAP8 | BPGAP1 | FLJ20185 | PP610 | Rho GTPase activating protein 8 | BPGAP1 interacts with RhoA. | BIND | 12944407 |
| ARHGDIA | GDIA1 | MGC117248 | RHOGDI | RHOGDI-1 | Rho GDP dissociation inhibitor (GDI) alpha | - | HPRD,BioGRID | 11149925 |
| ARHGDIB | D4 | GDIA2 | GDID4 | LYGDI | Ly-GDI | RAP1GN1 | RhoGDI2 | Rho GDP dissociation inhibitor (GDI) beta | - | HPRD,BioGRID | 9799233 |
| ARHGDIG | RHOGDI-3 | Rho GDP dissociation inhibitor (GDI) gamma | - | HPRD,BioGRID | 9113980 |
| ARHGEF1 | GEF1 | LBCL2 | LSC | P115-RHOGEF | SUB1.5 | Rho guanine nucleotide exchange factor (GEF) 1 | Biochemical Activity | BioGRID | 12748184 |
| ARHGEF11 | DKFZp667F1223 | GTRAP48 | KIAA0380 | PDZ-RHOGEF | Rho guanine nucleotide exchange factor (GEF) 11 | - | HPRD,BioGRID | 10526156 |
| ARHGEF12 | DKFZp686O2372 | KIAA0382 | LARG | PRO2792 | Rho guanine nucleotide exchange factor (GEF) 12 | - | HPRD,BioGRID | 11373293 |
| ARHGEF18 | KIAA0521 | MGC15913 | rho/rac guanine nucleotide exchange factor (GEF) 18 | - | HPRD,BioGRID | 11085924 |
| ARHGEF2 | DKFZp547L106 | DKFZp547P1516 | GEF | GEF-H1 | GEFH1 | KIAA0651 | LFP40 | P40 | rho/rac guanine nucleotide exchange factor (GEF) 2 | - | HPRD,BioGRID | 9857026 |
| ARHGEF3 | DKFZp434F2429 | FLJ98126 | GEF3 | MGC118905 | STA3 | XPLN | Rho guanine nucleotide exchange factor (GEF) 3 | - | HPRD,BioGRID | 12221096 |
| ARHGEF4 | ASEF | ASEF1 | GEF4 | STM6 | Rho guanine nucleotide exchange factor (GEF) 4 | - | HPRD,BioGRID | 10947987 |
| BID | FP497 | MGC15319 | MGC42355 | BH3 interacting domain death agonist | Co-purification | BioGRID | 15659383 |
| CASP10 | ALPS2 | FLICE2 | MCH4 | caspase 10, apoptosis-related cysteine peptidase | Co-purification | BioGRID | 15659383 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | Co-purification | BioGRID | 15659383 |
| CIT | CRIK | KIAA0949 | STK21 | citron (rho-interacting, serine/threonine kinase 21) | Far Western Reconstituted Complex Two-hybrid | BioGRID | 8543060 |12773565 |
| CNKSR1 | CNK | CNK1 | KSR | connector enhancer of kinase suppressor of Ras 1 | - | HPRD | 14749388 |
| CNKSR1 | CNK | CNK1 | KSR | connector enhancer of kinase suppressor of Ras 1 | hCNK1 interacts with RhoA. This interaction was modelled on a demonstrated interaction between human CNK1 and RhoA from an unspecified species. | BIND | 14749388 |
| CNTNAP1 | CASPR | CNTNAP | NRXN4 | P190 | contactin associated protein 1 | - | HPRD | 9407060 |
| CRMP1 | DPYSL1 | DRP-1 | DRP1 | collapsin response mediator protein 1 | Phenotypic Suppression | BioGRID | 12482610 |
| DAAM1 | FLJ41657 | KIAA0666 | dishevelled associated activator of morphogenesis 1 | - | HPRD | 11779461 |
| DEF6 | IBP | SLAT | differentially expressed in FDCP 6 homolog (mouse) | - | HPRD,BioGRID | 15023524 |
| DEF6 | IBP | SLAT | differentially expressed in FDCP 6 homolog (mouse) | DEF6 interacts with RhoA. This interaction was modelled on a demonstrated interaction between human DEF6 and RhoA from an unspecified species. | BIND | 15023524 |
| DGKQ | DAGK | DAGK4 | DAGK7 | diacylglycerol kinase, theta 110kDa | - | HPRD,BioGRID | 10066731 |
| DIAPH1 | DFNA1 | DIA1 | DRF1 | FLJ25265 | LFHL1 | hDIA1 | diaphanous homolog 1 (Drosophila) | Reconstituted Complex Two-hybrid | BioGRID | 12773565 |
| DOCK7 | KIAA1771 | ZIR2 | dedicator of cytokinesis 7 | - | HPRD | 12432077 |
| DPYSL2 | CRMP2 | DHPRP2 | DRP-2 | DRP2 | dihydropyrimidinase-like 2 | Phenotypic Suppression | BioGRID | 12482610 |
| EZR | CVIL | CVL | DKFZp762H157 | FLJ26216 | MGC1584 | VIL2 | ezrin | Co-purification | BioGRID | 15659383 |
| FADD | GIG3 | MGC8528 | MORT1 | Fas (TNFRSF6)-associated via death domain | Co-purification | BioGRID | 15659383 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | Co-purification | BioGRID | 15659383 |
| FASLG | APT1LG1 | CD178 | CD95L | FASL | TNFSF6 | Fas ligand (TNF superfamily, member 6) | Co-purification | BioGRID | 15659383 |
| FLNA | ABP-280 | ABPX | DKFZp434P031 | FLN | FLN1 | FMD | MNS | NHBP | OPD | OPD1 | OPD2 | filamin A, alpha (actin binding protein 280) | - | HPRD | 10051605 |
| GEFT | p63RhoGEF | RhoA/RAC/CDC42 exchange factor | - | HPRD,BioGRID | 15069594 |
| GMIP | - | GEM interacting protein | - | HPRD | 12093360 |
| GNA12 | MGC104623 | MGC99644 | NNX3 | RMP | gep | guanine nucleotide binding protein (G protein) alpha 12 | Reconstituted Complex | BioGRID | 12515866 |
| GRLF1 | GRF-1 | KIAA1722 | MGC10745 | P190-A | P190A | p190RhoGAP | glucocorticoid receptor DNA binding factor 1 | Reconstituted Complex | BioGRID | 9548756 |
| HSPA1A | FLJ54303 | FLJ54370 | FLJ54392 | FLJ54408 | FLJ75127 | HSP70-1 | HSP70-1A | HSP70I | HSP72 | HSPA1 | HSPA1B | heat shock 70kDa protein 1A | Affinity Capture-Western | BioGRID | 12490434 |
| ICMT | HSTE14 | MGC39955 | MST098 | MSTP098 | PCCMT | PCMT | PPMT | isoprenylcysteine carboxyl methyltransferase | - | HPRD,BioGRID | 9614111 |
| ITPR1 | INSP3R1 | IP3R | IP3R1 | SCA15 | SCA16 | inositol 1,4,5-triphosphate receptor, type 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12766172 |
| KCNA2 | HBK5 | HK4 | HUKIV | KV1.2 | MGC50217 | MK2 | NGK1 | RBK2 | potassium voltage-gated channel, shaker-related subfamily, member 2 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9635436 |
| KTN1 | CG1 | KIAA0004 | KNT | MGC133337 | MU-RMS-40.19 | kinectin 1 (kinesin receptor) | - | HPRD | 8769096 |11689693 |
| KTN1 | CG1 | KIAA0004 | KNT | MGC133337 | MU-RMS-40.19 | kinectin 1 (kinesin receptor) | Two-hybrid | BioGRID | 8769096 |11162552 |11689693 |12773565 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | Co-purification | BioGRID | 15659383 |
| MCF2 | DBL | MCF.2 cell line derived transforming sequence | - | HPRD,BioGRID | 10854437 |
| MCF2L | ARHGEF14 | DBS | FLJ12122 | KIAA0362 | OST | MCF.2 cell line derived transforming sequence-like | RhoA specifically interacts with Dbl-homology and pleckstrin-homology domains of the nucleotide exchange factor Dbs. This interaction is modelled on a demonstrated interaction between mouse Dbs and human RhoA. | BIND | 12006984 |
| MCF2L | ARHGEF14 | DBS | FLJ12122 | KIAA0362 | OST | MCF.2 cell line derived transforming sequence-like | - | HPRD,BioGRID | 12006984 |
| MPRIP | KIAA0864 | M-RIP | RHOIP3 | p116Rip | myosin phosphatase Rho interacting protein | - | HPRD | 14506264 |
| MPRIP | KIAA0864 | M-RIP | RHOIP3 | p116Rip | myosin phosphatase Rho interacting protein | - | HPRD,BioGRID | 9199174 |
| MSN | - | moesin | Co-purification | BioGRID | 15659383 |
| NET1 | ARHGEF8 | NET1A | neuroepithelial cell transforming 1 | Two-hybrid | BioGRID | 9535835 |
| OPHN1 | MRX60 | OPN1 | oligophrenin 1 | - | HPRD,BioGRID | 9582072 |11998687 |
| PDE6D | PDED | phosphodiesterase 6D, cGMP-specific, rod, delta | - | HPRD,BioGRID | 11786539 |
| PITPNM1 | DRES9 | FLJ44997 | NIR2 | PITPNM | RDGB | RDGB1 | RDGBA | RDGBA1 | Rd9 | phosphatidylinositol transfer protein, membrane-associated 1 | Nir2 interacts with RhoA. | BIND | 15125835 |
| PKN1 | DBK | MGC46204 | PAK1 | PKN | PKN-ALPHA | PRK1 | PRKCL1 | protein kinase N1 | - | HPRD,BioGRID | 9446575 |
| PKN2 | MGC150606 | MGC71074 | PAK2 | PRK2 | PRKCL2 | PRO2042 | Pak-2 | protein kinase N2 | - | HPRD,BioGRID | 8910519 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | - | HPRD,BioGRID | 12071848 |
| PLD1 | - | phospholipase D1, phosphatidylcholine-specific | - | HPRD,BioGRID | 11311142 |12593858 |
| PLEKHG2 | CLG | DKFZp667J2325 | FLJ00018 | FLJ22458 | FLJ38638 | pleckstrin homology domain containing, family G (with RhoGef domain) member 2 | - | HPRD,BioGRID | 11839748 |
| PPP1R12A | MBS | MGC133042 | MYPT1 | protein phosphatase 1, regulatory (inhibitor) subunit 12A | - | HPRD | 9354661 |
| PRKCA | AAG6 | MGC129900 | MGC129901 | PKC-alpha | PKCA | PRKACA | protein kinase C, alpha | in vivo | BioGRID | 11284700 |
| RABAC1 | PRA1 | PRAF1 | YIP3 | Rab acceptor 1 (prenylated) | - | HPRD | 11335720 |
| RAP1GDS1 | GDS1 | MGC118859 | MGC118861 | SmgGDS | RAP1, GTP-GDP dissociation stimulator 1 | - | HPRD,BioGRID | 11948427 |
| RHPN1 | KIAA1929 | ODF5 | RHOPHILIN | RHPN | rhophilin, Rho GTPase binding protein 1 | - | HPRD | 12221077 |
| RHPN2 | RhoBP | p76RBE | rhophilin, Rho GTPase binding protein 2 | - | HPRD | 12221077 |
| RHPN2 | RhoBP | p76RBE | rhophilin, Rho GTPase binding protein 2 | Two-hybrid | BioGRID | 12773565 |
| RICS | GC-GAP | GRIT | KIAA0712 | MGC1892 | p200RhoGAP | p250GAP | Rho GTPase-activating protein | Biochemical Activity | BioGRID | 12446789 |12454018 |12857875 |
| RIPK4 | ANKK2 | ANKRD3 | DIK | MGC129992 | MGC129993 | PKK | RIP4 | receptor-interacting serine-threonine kinase 4 | Two-hybrid | BioGRID | 9199174 |
| ROCK1 | MGC131603 | MGC43611 | P160ROCK | PRO0435 | Rho-associated, coiled-coil containing protein kinase 1 | - | HPRD,BioGRID | 8798490 |
| ROCK2 | KIAA0619 | Rho-associated, coiled-coil containing protein kinase 2 | Affinity Capture-Western | BioGRID | 11350930 |
| RTKN | - | rhotekin | RhoA interacts with RTKN (Rhotekin). This interaction was modeled on a demonstrated interaction between RhoA and RTKN from an unspecified species. | BIND | 15652748 |
| RTKN | - | rhotekin | Rhotekin interacts with RhoA-GTP. This interaction was modeled on a demonstrated interaction between Rhotekin from an unspecified species and human Rho-GTP. | BIND | 15467718 |
| RTKN | - | rhotekin | - | HPRD | 8662891 |
| RTKN | - | rhotekin | RTKN (rhotekin) interacts with RhoA. This interaction was modeled on a demonstrated interaction between mouse RTKN and human RhoA. | BIND | 15766663 |
| RTKN | - | rhotekin | Reconstituted Complex | BioGRID | 12593858 |
| RTKN | - | rhotekin | RhoA interacts with Rhotekin. This interaction was modeled on a demonstrated interaction between human RhoA and Rhotekin from an unspecified species. | BIND | 15834426 |
| RTKN | - | rhotekin | Active, GTP-bound RhoA interacts with the RTKN (Rhotekin) Rho binding domain. This interaction was modelled on a demonstrated interaction between human RhoA and Rhotekin from an unspecified species. | BIND | 15889147 |
| SH3BP1 | - | SH3-domain binding protein 1 | - | HPRD | 9548756 |
| SPRED2 | FLJ21897 | FLJ31917 | MGC163164 | Spred-2 | sprouty-related, EVH1 domain containing 2 | - | HPRD | 15184877 |
| SRGAP1 | ARHGAP13 | FLJ22166 | KIAA1304 | SLIT-ROBO Rho GTPase activating protein 1 | - | HPRD | 11672528 |
| TEC | MGC126760 | MGC126762 | PSCTK4 | tec protein tyrosine kinase | Reconstituted Complex | BioGRID | 12515866 |
| TGM2 | G-ALPHA-h | GNAH | TG2 | TGC | transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) | - | HPRD | 11350930 |
| TNFRSF10B | CD262 | DR5 | KILLER | KILLER/DR5 | TRAIL-R2 | TRAILR2 | TRICK2 | TRICK2A | TRICK2B | TRICKB | ZTNFR9 | tumor necrosis factor receptor superfamily, member 10b | Co-purification | BioGRID | 15659383 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | Co-purification | BioGRID | 15659383 |
| TRIO | FLJ42780 | tgat | triple functional domain (PTPRF interacting) | - | HPRD,BioGRID | 10948190 |
| TRPC1 | HTRP-1 | MGC133334 | MGC133335 | TRP1 | transient receptor potential cation channel, subfamily C, member 1 | - | HPRD,BioGRID | 12766172 |
| VAV2 | - | vav 2 guanine nucleotide exchange factor | - | HPRD | 11909943 |
| VAV3 | FLJ40431 | vav 3 guanine nucleotide exchange factor | - | HPRD,BioGRID | 10523675 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION | 115 | 81 | All SZGR 2.0 genes in this pathway |
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ADHERENS JUNCTION | 75 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG PATHOGENIC ESCHERICHIA COLI INFECTION | 59 | 36 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG COLORECTAL CANCER | 62 | 47 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CCR3 PATHWAY | 23 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ECM PATHWAY | 24 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RACCYCD PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INTEGRIN PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EDG1 PATHWAY | 27 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AKAPCENTROSOME PATHWAY | 15 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RAS PATHWAY | 23 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RHO PATHWAY | 32 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AKAP13 PATHWAY | 12 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CARDIACEGF PATHWAY | 18 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAL PATHWAY | 19 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CDC42RAC PATHWAY | 16 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PAR1 PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TFF PATHWAY | 21 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA UCALPAIN PATHWAY | 18 | 11 | All SZGR 2.0 genes in this pathway |
| SA TRKA RECEPTOR | 17 | 15 | All SZGR 2.0 genes in this pathway |
| PID ENDOTHELIN PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| PID PRL SIGNALING EVENTS PATHWAY | 23 | 17 | All SZGR 2.0 genes in this pathway |
| PID RHOA PATHWAY | 45 | 33 | All SZGR 2.0 genes in this pathway |
| PID AURORA B PATHWAY | 39 | 24 | All SZGR 2.0 genes in this pathway |
| PID LYSOPHOSPHOLIPID PATHWAY | 66 | 53 | All SZGR 2.0 genes in this pathway |
| PID WNT NONCANONICAL PATHWAY | 32 | 26 | All SZGR 2.0 genes in this pathway |
| PID ER NONGENOMIC PATHWAY | 41 | 35 | All SZGR 2.0 genes in this pathway |
| PID MET PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID PTP1B PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P3 PATHWAY | 29 | 24 | All SZGR 2.0 genes in this pathway |
| PID AVB3 OPN PATHWAY | 31 | 29 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P4 PATHWAY | 14 | 11 | All SZGR 2.0 genes in this pathway |
| PID RHOA REG PATHWAY | 46 | 30 | All SZGR 2.0 genes in this pathway |
| PID RET PATHWAY | 39 | 29 | All SZGR 2.0 genes in this pathway |
| PID TXA2PATHWAY | 57 | 43 | All SZGR 2.0 genes in this pathway |
| PID SHP2 PATHWAY | 58 | 46 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P1 PATHWAY | 21 | 18 | All SZGR 2.0 genes in this pathway |
| PID NETRIN PATHWAY | 32 | 27 | All SZGR 2.0 genes in this pathway |
| PID ARF6 DOWNSTREAM PATHWAY | 15 | 14 | All SZGR 2.0 genes in this pathway |
| PID MTOR 4PATHWAY | 69 | 55 | All SZGR 2.0 genes in this pathway |
| PID IL2 1PATHWAY | 55 | 43 | All SZGR 2.0 genes in this pathway |
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| PID PLK1 PATHWAY | 46 | 25 | All SZGR 2.0 genes in this pathway |
| PID THROMBIN PAR4 PATHWAY | 15 | 11 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI PATHWAY | 49 | 40 | All SZGR 2.0 genes in this pathway |
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN NASCENT AJ PATHWAY | 39 | 33 | All SZGR 2.0 genes in this pathway |
| PID AMB2 NEUTROPHILS PATHWAY | 41 | 32 | All SZGR 2.0 genes in this pathway |
| PID AVB3 INTEGRIN PATHWAY | 75 | 53 | All SZGR 2.0 genes in this pathway |
| PID LIS1 PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 4 PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| PID EPHA FWDPATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN KERATINOCYTE PATHWAY | 21 | 19 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID TRKR PATHWAY | 62 | 48 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN STABILIZATION PATHWAY | 42 | 34 | All SZGR 2.0 genes in this pathway |
| PID VEGFR1 2 PATHWAY | 69 | 57 | All SZGR 2.0 genes in this pathway |
| PID THROMBIN PAR1 PATHWAY | 43 | 32 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 2 PATHWAY | 33 | 27 | All SZGR 2.0 genes in this pathway |
| PID TOLL ENDOGENOUS PATHWAY | 25 | 19 | All SZGR 2.0 genes in this pathway |
| PID NCADHERIN PATHWAY | 33 | 32 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P2 PATHWAY | 24 | 19 | All SZGR 2.0 genes in this pathway |
| PID EPHA2 FWD PATHWAY | 19 | 16 | All SZGR 2.0 genes in this pathway |
| PID FAK PATHWAY | 59 | 45 | All SZGR 2.0 genes in this pathway |
| PID TGFBR PATHWAY | 55 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY RHO GTPASES | 113 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | 16 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K AKT ACTIVATION | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | 81 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME GPVI MEDIATED ACTIVATION CASCADE | 31 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA1213 SIGNALLING EVENTS | 74 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMA4D IN SEMAPHORIN SIGNALING | 32 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMAPHORIN INTERACTIONS | 68 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | 27 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | 25 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME G PROTEIN BETA GAMMA SIGNALLING | 28 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | 63 | 42 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 DN | 153 | 100 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| LI LUNG CANCER | 41 | 30 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| ALCALA APOPTOSIS | 88 | 60 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| ZUCCHI METASTASIS DN | 44 | 29 | All SZGR 2.0 genes in this pathway |
| IIZUKA LIVER CANCER PROGRESSION L0 L1 DN | 21 | 14 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS UP | 69 | 41 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA | 50 | 31 | All SZGR 2.0 genes in this pathway |
| MACLACHLAN BRCA1 TARGETS UP | 21 | 16 | All SZGR 2.0 genes in this pathway |
| ZHANG PROLIFERATING VS QUIESCENT | 51 | 41 | All SZGR 2.0 genes in this pathway |
| DELLA RESPONSE TO TSA AND BUTYRATE | 21 | 17 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA EARLY RESPONSE TO TGFB1 | 58 | 43 | All SZGR 2.0 genes in this pathway |
| RAMPON ENRICHED LEARNING ENVIRONMENT EARLY UP | 15 | 12 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C3 | 14 | 10 | All SZGR 2.0 genes in this pathway |
| JOSEPH RESPONSE TO SODIUM BUTYRATE UP | 31 | 21 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND FIBROBLAST | 132 | 93 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 9 11 TRANSLOCATION | 130 | 87 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| ABDELMOHSEN ELAVL4 TARGETS | 16 | 13 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN DAUNORUBICIN B ALL DN | 12 | 10 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-142-5p | 330 | 337 | 1A,m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-185 | 987 | 994 | 1A,m8 | hsa-miR-185brain | UGGAGAGAAAGGCAGUUC |
| miR-200bc/429 | 133 | 139 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-361 | 742 | 749 | 1A,m8 | hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC |
| miR-381 | 614 | 620 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-485-3p | 62 | 68 | m8 | hsa-miR-485-3p | GUCAUACACGGCUCUCCUCUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.