| PID ECADHERIN STABILIZATION PATHWAY
| 42 | 34 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP
| 579 | 346 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS UP
| 306 | 188 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP
| 133 | 78 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP
| 430 | 232 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED UP
| 78 | 48 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP
| 722 | 443 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN
| 508 | 354 | All SZGR 2.0 genes in this pathway |
| BERENJENO ROCK SIGNALING NOT VIA RHOA DN
| 48 | 34 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN
| 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 7
| 8 | 6 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM UP
| 176 | 111 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN
| 514 | 319 | All SZGR 2.0 genes in this pathway |
| KIM MYC AMPLIFICATION TARGETS DN
| 97 | 51 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN
| 848 | 527 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN
| 514 | 330 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN
| 411 | 249 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES
| 648 | 385 | All SZGR 2.0 genes in this pathway |
| MORI LARGE PRE BII LYMPHOCYTE DN
| 58 | 39 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE UP
| 90 | 62 | All SZGR 2.0 genes in this pathway |
| KIM GERMINAL CENTER T HELPER UP
| 66 | 42 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN
| 50 | 32 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN
| 408 | 274 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM
| 302 | 191 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN
| 830 | 547 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS UP
| 175 | 116 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR DN
| 101 | 70 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A DN
| 76 | 57 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP
| 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN
| 504 | 323 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP
| 262 | 186 | All SZGR 2.0 genes in this pathway |
| KEEN RESPONSE TO ROSIGLITAZONE DN
| 106 | 68 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN
| 318 | 220 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP
| 280 | 183 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS DN
| 314 | 188 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP
| 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP
| 783 | 442 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 2G
| 171 | 96 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP
| 323 | 240 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP
| 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN
| 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN
| 245 | 150 | All SZGR 2.0 genes in this pathway |
| LEE RECENT THYMIC EMIGRANT
| 227 | 128 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SURVIVAL UP
| 73 | 49 | All SZGR 2.0 genes in this pathway |
| GUTIERREZ CHRONIC LYMPHOCYTIC LEUKEMIA DN
| 56 | 39 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN
| 225 | 124 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE
| 721 | 492 | All SZGR 2.0 genes in this pathway |
| ZEMBUTSU SENSITIVITY TO VINBLASTINE
| 18 | 12 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 M
| 216 | 124 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT
| 572 | 352 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT
| 567 | 365 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP
| 504 | 321 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN
| 315 | 215 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN
| 553 | 343 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP
| 344 | 215 | All SZGR 2.0 genes in this pathway |
| GUO TARGETS OF IRS1 AND IRS2
| 98 | 67 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1
| 467 | 251 | All SZGR 2.0 genes in this pathway |
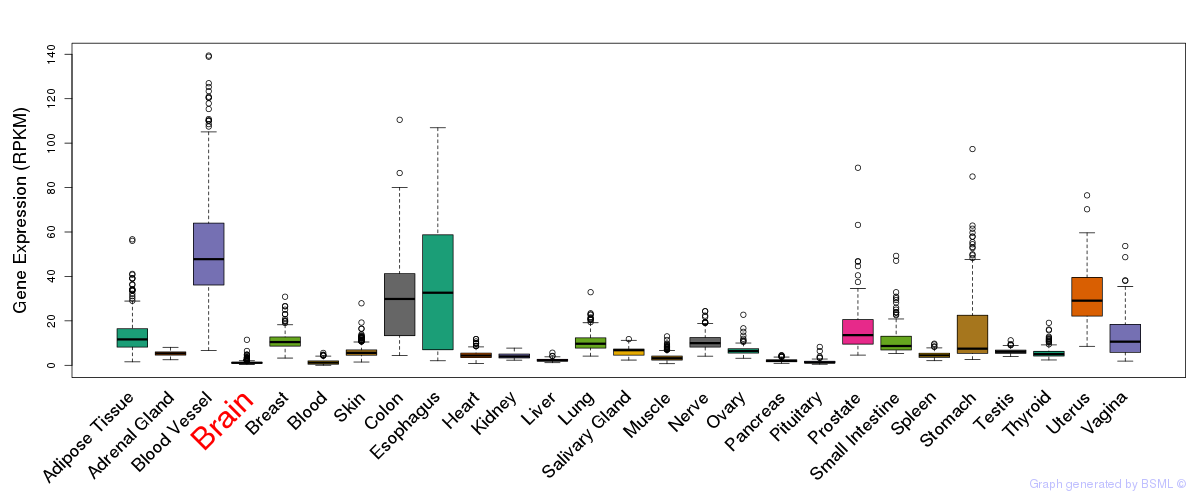
 CV:PheWAS
CV:PheWAS eQTL annotation
eQTL annotation